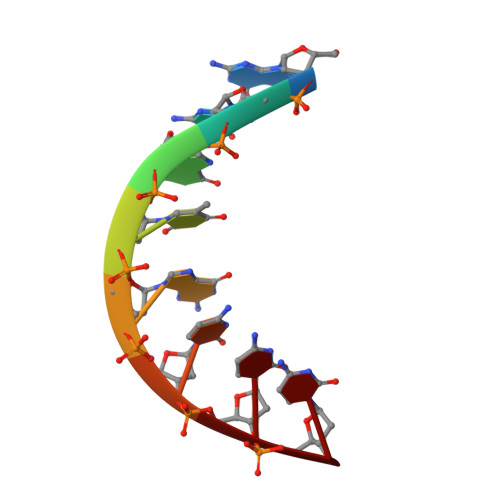
Structures of the mismatched duplex d(GGGTGCCC) and one of its Watson-Crick analogues d(GGGCGCCC).
Rabinovich, D., Haran, T., Eisenstein, M., Shakked, Z.(1988) J Mol Biol 200: 151-161
- PubMed: 3379638
- DOI: https://doi.org/10.1016/0022-2836(88)90340-3
- Primary Citation of Related Structures:
1VT7 - PubMed Abstract:
The mismatched duplex d(GGGTGCCC) (I) and its two Watson-Crick analogues (dGGGCGCCC) (II) and d(GGGTACCC) (III) were synthesized. The X-ray crystal structures of (I) and (II) were determined at resolutions of 2.5 and 1.7 A (1 A = 0.1 nm) and refined to R factors of 15 and 16%, respectively. (I) and (II) crystallize as A-DNA doublehelical octamers in space groups P61 and P4(3)2(1)2, respectively, and are stable at room temperature. The central two G.T mispairs of (I) adopt the wobble geometry as observed in other G.T mismatches. The two structures differ significantly in their local conformational features at the central helical regions as well as in some global ones. In particular, T-G adopts a large helical twist (44 degrees) whereas C-G adopts a small one (24 degrees). This difference can be rationalized on the basis of simple geometrical considerations. Base-pair stacking energies which were calculated for the two duplexes indicate that (I) is destabilized with respect to (II). Helix-coil transition measurements were performed for each of the three oligomers by means of ultraviolet absorbance spectrophotometry. The results indicate that the stability of the duplexes and the co-operativity of the transition are in the following order: (I) less than (III) less than (II). Such studies may help in understanding why certain regions of DNA are more likely to undergo spontaneous mutations than others.
Organizational Affiliation:
Department of Structural Chemistry, Weizmann Institute of Science Rehovot, Israel.