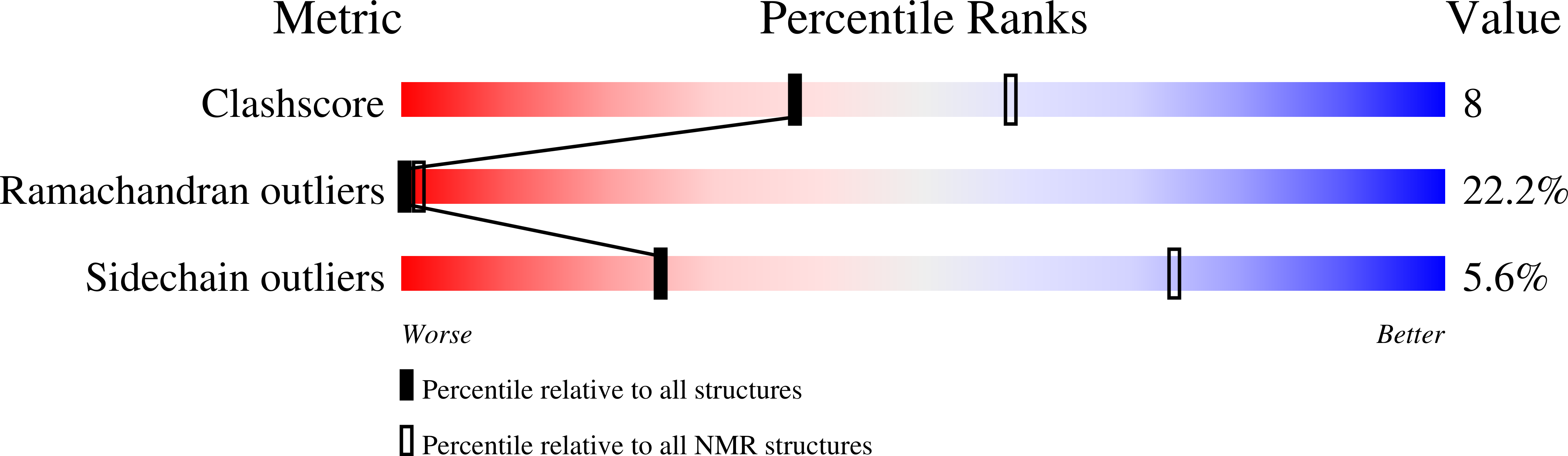Design of a New Mimochrome with Unique Topology.
Lombardi, A., Nastri, F., Marasco, D., Maglio, O., De Sanctis, G., Sinibaldi, F., Santucci, R., Coletta, M., Pavone, V.(2003) Chemistry 9: 5643-5654
- PubMed: 14639648
- DOI: https://doi.org/10.1002/chem.200304831
- Primary Citation of Related Structures:
1VL3 - PubMed Abstract:
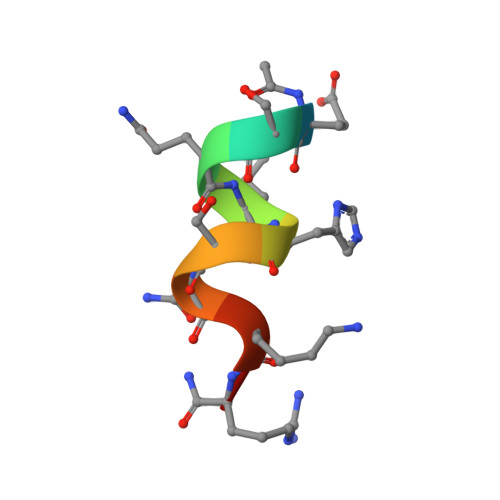
Peptide-based metalloprotein models represent useful systems to help understand how metalloproteins can support different functions, by the use of similar metal ion cofactors. In order to shed light on the role of the protein matrix in modulating the heme properties, we developed new models: mimochromes. They are pseudo-C(2) symmetric systems, composed of two helical peptides covalently linked to the deuteroporphyrin. The use of C(2) symmetry is particularly advantageous, because it simplifies the design, synthesis and characterization. However, it leaves the problem of possible diastereomeric forms. In the cobalt complex of the first derivative, mimochrome I, Lambda and Delta isomers were indeed experimentally observed. All the insights derived from the Co(III)-mimochrome I structure were used to obtain a re-designed molecule, mimochrome IV. The spectroscopic characterization of the iron and cobalt derivatives suggested the presence of the Lambda isomer as unique species. The NMR solution structure of the diamagnetic Co(III)-mimochrome IV confirmed the ability of the molecule to adopt a unique topology, and revealed the peptide chains to be in helical conformation, as designed. The insertion of intramolecular, inter-chain interactions was successful in favoring the formation of one of the two possible diastereomers. The stereochemically stable structure of mimochrome IV provides an attractive model for modulating the redox potential of the heme, by simple changing the peptide chain composition around the heme.
Organizational Affiliation:
Department of Chemistry, University of Naples Federico II, Complesso Universitario Monte S. Angelo, Via Cynthia, 80126 Naples, Italy.