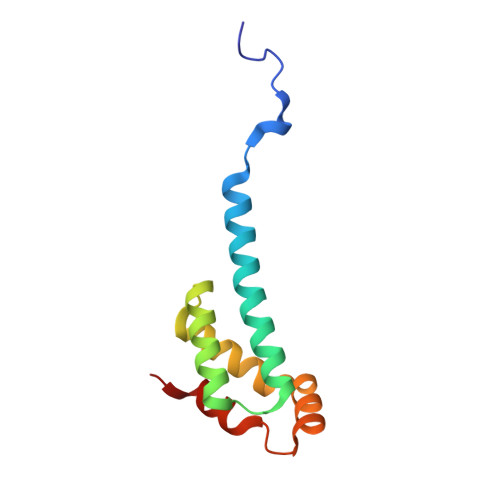
Crystal Structure of the Ent Domain of Human Emsy.
Chavali, G.B., Ekblad, C.M., Basu, B.P., Brissett, N.C., Veprintsev, D., Hughes-Davies, L., Kouzarides, T., Itzhaki, L.S., Doherty, A.J.(2005) J Mol Biol 350: 964
- PubMed: 15978617
- DOI: https://doi.org/10.1016/j.jmb.2005.05.047
- Primary Citation of Related Structures:
1UZ3 - PubMed Abstract:
EMSY is a recently discovered gene encoding a BRCA2-associated protein and is amplified in some sporadic breast and ovarian cancers. The EMSY sequence contains no known domain except for a conserved approximately 100 residue segment at the N terminus. This so-called ENT domain is unique in the human genome, although multiple copies are found in Arabidopsis proteins containing members of the Royal family of chromatin remodelling domains. Here, we report the crystal structure of the ENT domain of EMSY, consisting of a unique arrangement of five alpha-helices that fold into a helical bundle arrangement. The fold shares regions of structural homology with the DNA-binding domain of homeodomain proteins. The ENT domain forms a homodimer via the anti-parallel packing of the extended N-terminal alpha-helix of each molecule. It is stabilized mainly by hydrophobic residues at the dimer interface and has a dissociation constant in the low micromolar range. The dimerisation of EMSY mediated by the ENT domain could provide flexibility for it to bind two or more different substrates simultaneously.
Organizational Affiliation:
Department of Haematology, Cambridge Institute for Medical Research, University of Cambridge, Hills Rd, Cambridge CB2 2XY, UK.