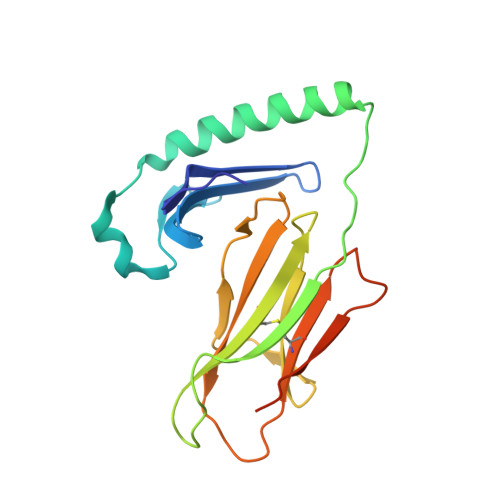
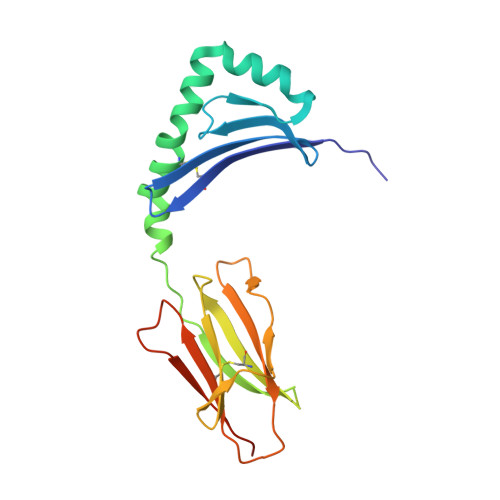
Crystal Structure of Hla-Dq0602 that Protects Against Type 1 Diabetes and Confers Strong Susceptibility to Narcolepsy
Siebold, C., Hansen, B.E., Wyer, J.R., Harlos, K., Esnouf, R.E., Svejgaard, A., Bell, J.I., Strominger, J.L., Jones, E.Y., Fugger, L.(2004) Proc Natl Acad Sci U S A 101: 1999
- PubMed: 14769912
- DOI: https://doi.org/10.1073/pnas.0308458100
- Primary Citation of Related Structures:
1UVQ - PubMed Abstract:
The MHC class II molecule DQ0602 confers strong susceptibility to narcolepsy but dominant protection against type 1 diabetes. The crystal structure of DQ0602 reveals the molecular features underlying these contrasting genetic properties. Structural comparisons to homologous DQ molecules with differential disease associations highlight a previously unrecognized interplay between the volume of the P6 pocket and the specificity of the P9 pocket, which implies that presentation of an expanded peptide repertoire is critical for dominant protection against type 1 diabetes. In narcolepsy, the volume of the P4 pocket appears central to the susceptibility, suggesting that the presentation of a specific peptide population plays a major role.
Organizational Affiliation:
Division of Structural Biology, The Henry Wellcome Building for Genomic Medicine, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, United Kingdom.