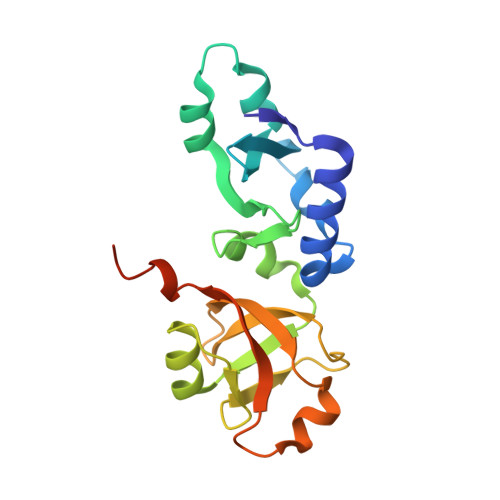
Crystal structure of KD93, a novel protein expressed in human hematopoietic stem/progenitor cells.
Liu, J.F., Wang, X.Q., Wang, Z.X., Chen, J.R., Jiang, T., An, X.M., Chang, W.R., Liang, D.C.(2004) J Struct Biol 148: 370-374
- PubMed: 15522784
- DOI: https://doi.org/10.1016/j.jsb.2004.06.010
- Primary Citation of Related Structures:
1SQW - PubMed Abstract:
The crystal structure of a novel hypothetical protein, KD93, expressed in human hematopoietic stem/progenitor cells, was determined at 1.9A resolution using the multiple-wavelength anomalous dispersion (MAD) method. The protein KD93, which is encoded by the open reading frame HSPC031, is a NIP7 homologue and belongs to the UPF0113 family. The structural and functional information for the group of homologues has not yet been determined. Crystallographic analysis revealed that the overall fold of KD93 consists of two interlinked alpha/beta domains. Structure-based homology analysis with DALI revealed that the C domain of KD93 matches the PUA domain of some RNA modification enzymes, especially that of archaeosine tRNA-ribosyltransferase (ArcTGT), which suggests that its possible molecular function is related to RNA binding. The difference between the RNA binding regions of KD93 and ArcTGT in amino acid constitution and surface electrostatic potential indicate that they may have different RNA binding modes. The N domain of KD93 is a unique structure with no obvious similarity to other proteins with known three-dimensional structures. The high-resolution structure of KD93 provides a first view of a member of the family of hypothetical proteins. And the structure provides a framework to deduce and assay the molecular function of other proteins of the UPF0113 family.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Institute of Biophysics, Academia Sinica, Beijing 100101, People's Republic of China.