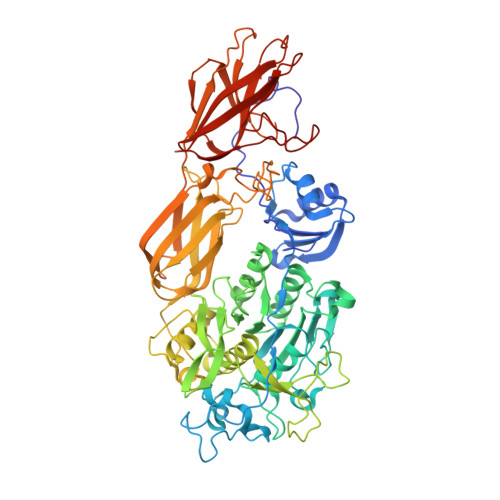
Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin.
Kim, J.S., Kluskens, L.D., de Vos, W.M., Huber, R., van der Oost, J.(2004) J Mol Biol 335: 787-797
- PubMed: 14687574
- DOI: https://doi.org/10.1016/j.jmb.2003.11.006
- Primary Citation of Related Structures:
1R6V - PubMed Abstract:
Structure-forming fibrous proteins like keratins, gelatins and collagens are degraded only by a few proteases as their tight packing limits access to the potential cleavage sites. To understand the keratin degradation in detail, we describe the first crystal structure of a keratin-degrading enzyme (keratinase), fervidolysin, from Fervidobacterium pennivorans as an immature form with propeptide (PD)-bound. The 1.7A resolution crystal structure shows that the protease is composed of four domains: a catalytic domain (CD), two beta-sandwich domains (SDs), and the PD domain. A structural alignment shows a distant relationship between the PD-CD substructure of fervidolysin and pro-subtilisin E. Tight binding of PD to the remaining part of the protease is mediated by hydrogen bonds along the domain surfaces and around the active cleft, and by the clamps to SD1 and SD2. The crystal structure of this multi-domain protein fervidolysin provides insights into proenzyme activation and the role of non-catalytic domains, suggesting a functional relationship to the fibronectin (FN)-like domains of the human promatrix metalloprotease-2 (proMMP-2) that degrades the fibrous polymeric substrate gelatin.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Abteilung Strukturforschung, Am Klopferspitz 18a, D-82152 Martinsried, Germany. jkim@biochem.mpg.de