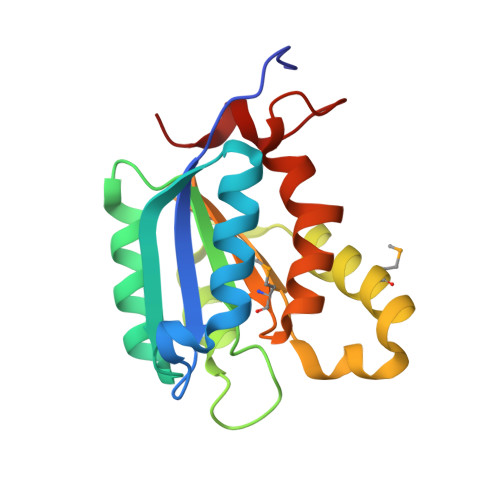
Structure of the molybdenum-cofactor biosynthesis protein MoaB of Escherichia coli.
Bader, G., Gomez-Ortiz, M., Haussmann, C., Bacher, A., Huber, R., Fischer, M.(2004) Acta Crystallogr D Biol Crystallogr 60: 1068-1075
- PubMed: 15159566
- DOI: https://doi.org/10.1107/S0907444904007164
- Primary Citation of Related Structures:
1R2K - PubMed Abstract:
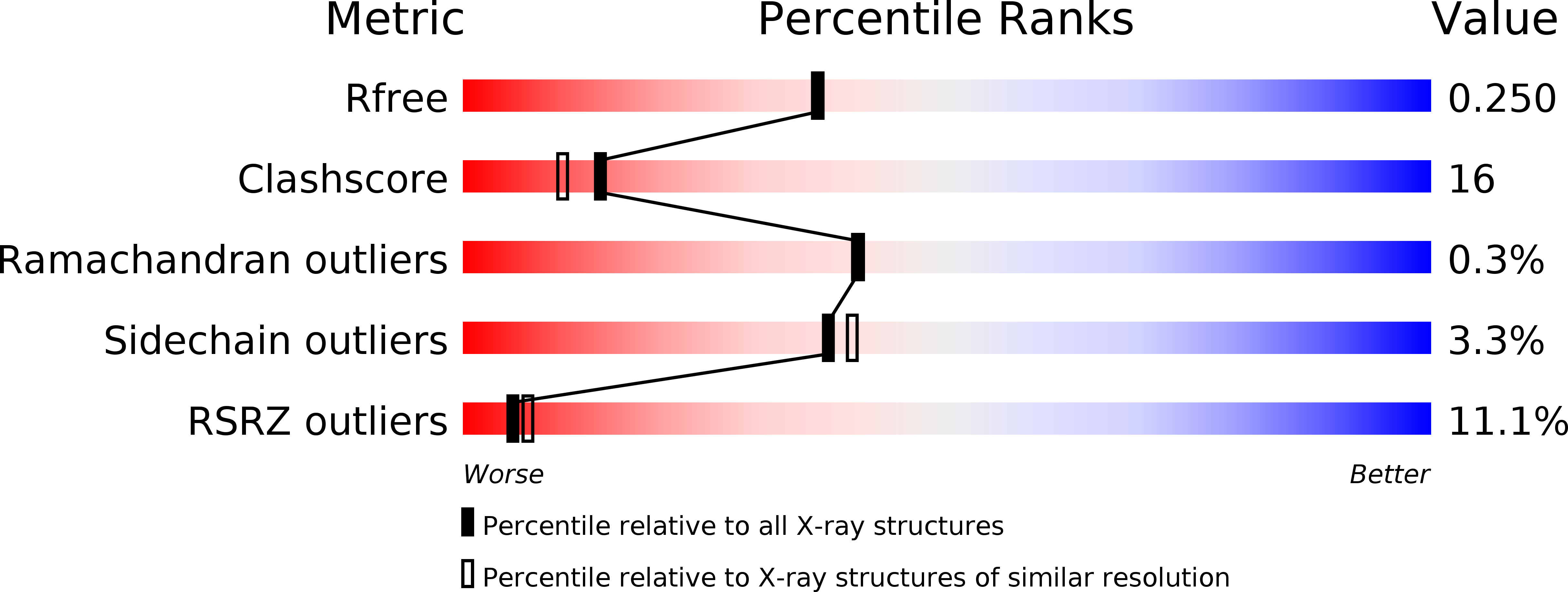
The moaABC operon of Escherichia coli is involved in early steps of the biosynthesis of the molybdenum-binding cofactor molybdopterin, but the precise functions of the cognate proteins are not known. The crystal structure of the MoaB protein from E. coli was determined by multiple anomalous dispersion at 2.1 angstroms A resolution and refined to an R factor of 20.4% (Rfree = 25.0%). The protein is a 32-symmetric hexamer, with the monomers consisting of a central beta-sheet flanked by helices on both sides. The overall fold of the monomer is similar to those of the MogA protein of E. coli, the G-domains of rat and human gephyrin and the G-domains of Cnx1 protein from A. thaliana, all of which are involved in the insertion of an unknown molybdenum species into molybdopterin to form the molybdenum cofactor. Furthermore, the MoaB protein shows significant sequence similarity to the cinnamon protein from Drosophila melanogaster. In addition to other functions, all these proteins are involved in the biosynthesis of the molybdenum cofactor and have been shown to bind molybdopterin. The close structural homology to MogA and the gephyrin and Cnx1 domains suggests that MoaB may bind a hitherto unidentified pterin compound, possibly an intermediate in molybdopterin biosynthesis.
Organizational Affiliation:
Abteilung Strukturforschung, Max-Planck-Institut für Biochemie, Am Klopferspitz 18a, D-82152 Martinsried, Germany. gerd.bader@vie.boehringer-ingelheim.com