Comparison of Kifunensine and 1-Deoxymannojirimycin Binding to Class I and II alpha-Mannosidases Demonstrates Different Saccharide Distortions in Inverting and Retaining Catalytic Mechanisms
Shah, N., Kuntz, D.A., Rose, D.R.(2003) Biochemistry 42: 13812-13816
- PubMed: 14636047
- DOI: https://doi.org/10.1021/bi034742r
- Primary Citation of Related Structures:
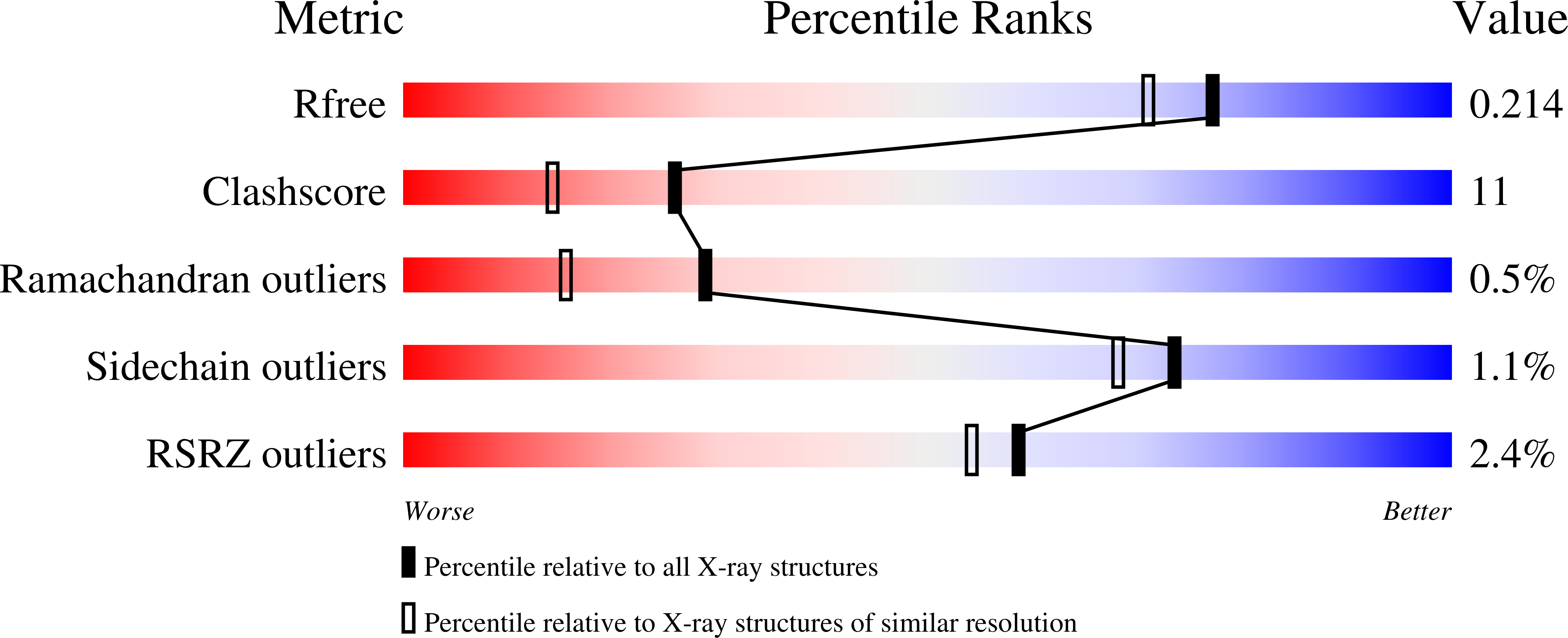
1PS3 - PubMed Abstract:
Mannosidases are key enzymes in the eukaryotic N-glycosylation pathway. These enzymes fall into two broad classes (I and II) and are characteristically different in catalytic mechanism, sequence, and structure. Kifunensine is an alkaloid that is a strong inhibitor against class I alpha-mannosidases but is only a weak inhibitor against class II alpha-mannosidases. In this paper, the 1.80 A resolution crystal structure of kifunensine bound to Drosophila melanogaster Golgi alpha-mannosidase II (dGMII) is presented. Kifunensine adopts a (1,4)B boat conformation in the class II dGMII, which contrasts the (1)C(4) chair conformation seen in class I human endoplasmic reticulum alpha1,2 mannosidase (hERMI, PDB ). The observed conformations are higher in conformational energy than the global minimum (4)C(1) conformation, although the conformation in hERMI is closer to the minimum, as supported by an energy calculation. Differing conformations of 1-deoxymannojirimycin were also observed: a (4)C(1) and (1)C(4) conformation in dGMII and hERMI, respectively. Thus, these two alpha-mannosidase classes distort these inhibitors in distinct manners. This is likely indicative of the binding characteristics of the two different catalytic mechanisms of these enzymes.
Organizational Affiliation:
Department of Medical Biophysics, University of Toronto, Ontario, Canada.