Structure of Viscotoxin A3: Disulfide Location from Weak Sad Data
Debreczeni, J.E., Girmann, B., Zeeck, A., Kratzner, R., Sheldrick, G.M.(2003) Acta Crystallogr D Biol Crystallogr 59: 2125
- PubMed: 14646070
- DOI: https://doi.org/10.1107/s0907444903018973
- Primary Citation of Related Structures:
1OKH - PubMed Abstract:
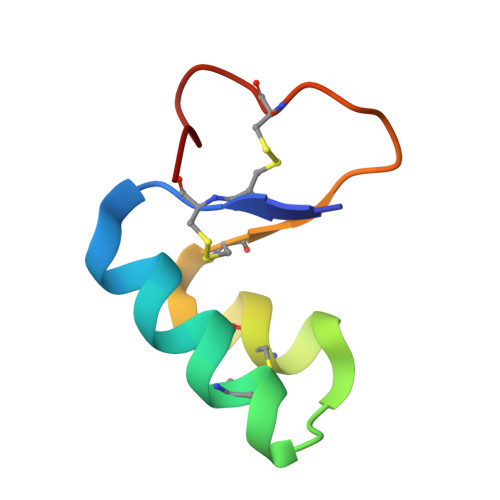
The crystal structure of viscotoxin A3 (VT A3) extracted from European mistletoe (Viscum album L.) has been solved using the anomalous diffraction of the native S atoms measured in-house with Cu Kalpha radiation to a resolution of 2.2 A and truncated to 2.5 A. A 1.75 A resolution synchrotron data set was used for phase expansion and refinement. An innovation in the dual-space substructure-solution program SHELXD enabled the individual S atoms of the disulfide bonds to be located using the Cu Kalpha data; this resulted in a marked improvement in the phasing compared with the use of super-S atoms. The VT A3 monomer consists of 46 amino acids with three disulfide bridges and has an overall fold resembling the canonical architecture of the alpha- and beta-thionins, a capital letter L. The asymmetric unit consists of two monomers related by a local twofold axis and held together by hydrophobic interactions between the monomer units. One phosphate anion (confirmed by 31P-NMR and MS) is associated with each monomer.
Organizational Affiliation:
Lehrstuhl für Strukturchemie, Georg-August Universität, Tammannstrasse 4, Göttingen, Germany.