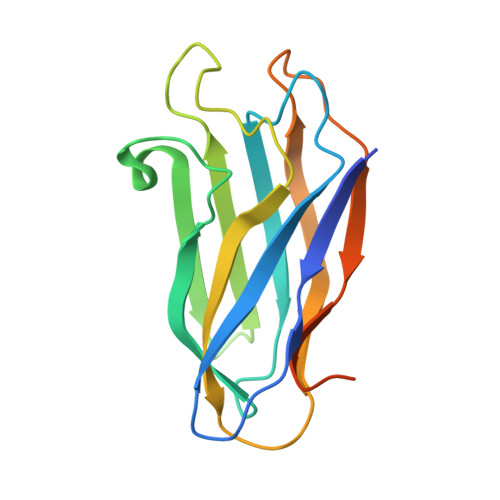
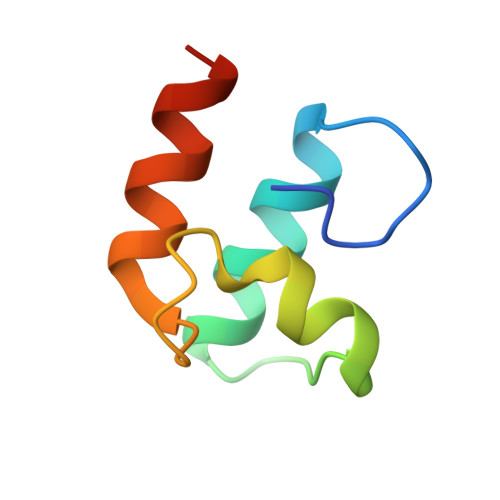
Cellulosome Assembly Revealed by the Crystal Structure of the Cohesin-Dockerin Complex
Carvalho, A.L., Dias, F.M.V., Prates, J.A.M., Ferreira, L.M.A., Gilbert, H.J., Davies, G.J., Romao, M.J., Fontes, C.M.G.A.(2003) Proc Natl Acad Sci U S A 100: 13809
- PubMed: 14623971
- DOI: https://doi.org/10.1073/pnas.1936124100
- Primary Citation of Related Structures:
1OHZ - PubMed Abstract:
The utilization of organized supramolecular assemblies to exploit the synergistic interactions afforded by close proximity, both for enzymatic synthesis and for the degradation of recalcitrant substrates, is an emerging theme in cellular biology. Anaerobic bacteria harness a multiprotein complex, termed the "cellulosome," for efficient degradation of the plant cell wall. This megadalton catalytic machine organizes an enzymatic consortium on a multifaceted molecular scaffold whose "cohesin" domains interact with corresponding "dockerin" domains of the enzymes. Here we report the structure of the cohesin-dockerin complex from Clostridium thermocellum at 2.2-A resolution. The data show that the beta-sheet cohesin domain interacts predominantly with one of the helices of the dockerin. Whereas the structure of the cohesin remains essentially unchanged, the loop-helix-helix-loop-helix motif of the dockerin undergoes conformational change and ordering compared with its solution structure, although the classical 12-residue EF-hand coordination to two calcium ions is maintained. Significantly, internal sequence duplication within the dockerin is manifested in near-perfect internal twofold symmetry, suggesting that both "halves" of the dockerin may interact with cohesins in a similar manner, thus providing a higher level of structure to the cellulosome and possibly explaining the presence of "polycellulosomes." The structure provides an explanation for the lack of cross-species recognition between cohesin-dockerin pairs and thus provides a blueprint for the rational design, construction, and exploitation of these catalytic assemblies.
Organizational Affiliation:
Rede de Química e Tecnologia/Centro de Química Fina e Biotecnologia (REQUIMTE/CQFB), Departamento de Química, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa, 2829-516 Caparica, Portugal.