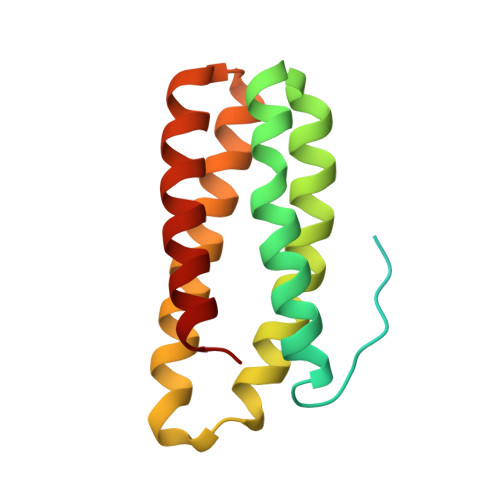
Crystal structure of the PsbQ protein of photosystem II from higher plants
Calderone, V., Trabucco, M., Vujicic, A., Battistutta, R., Giacometti, G.M., Andreucci, F., Barbato, R., Zanotti, G.(2003) EMBO Rep 4: 900-905
- PubMed: 12949587
- DOI: https://doi.org/10.1038/sj.embor.embor923
- Primary Citation of Related Structures:
1NZE - PubMed Abstract:
The smallest extrinsic polypeptide of the water-oxidizing complex (PsbQ) was extracted and purified from spinach (Spinacia oleracea) photosystem II (PSII) membranes. It was then crystallized in the presence of Zn(2+) and its structure was determined by X-ray diffraction at 1.95-A resolution using the multi-wavelength anomalous diffraction method, with the zinc as the anomalous scatterer. The crystal structure shows that the core of the protein is a four-helix bundle, whereas the amino-terminal portion, which possibly interacts with the photosystem core, is not visible in the crystal. The distribution of positive and negative charges on the protein surface might explain the ability of PsbQ to increase the binding of Cl(-) and Ca(2+) and make them available to PSII.
Organizational Affiliation:
Dipartimento di Chimica Organica e Centro Biopolimeri del CNR, Università di Padova, Via Marzolo 1, 35131 Padua, Italy.