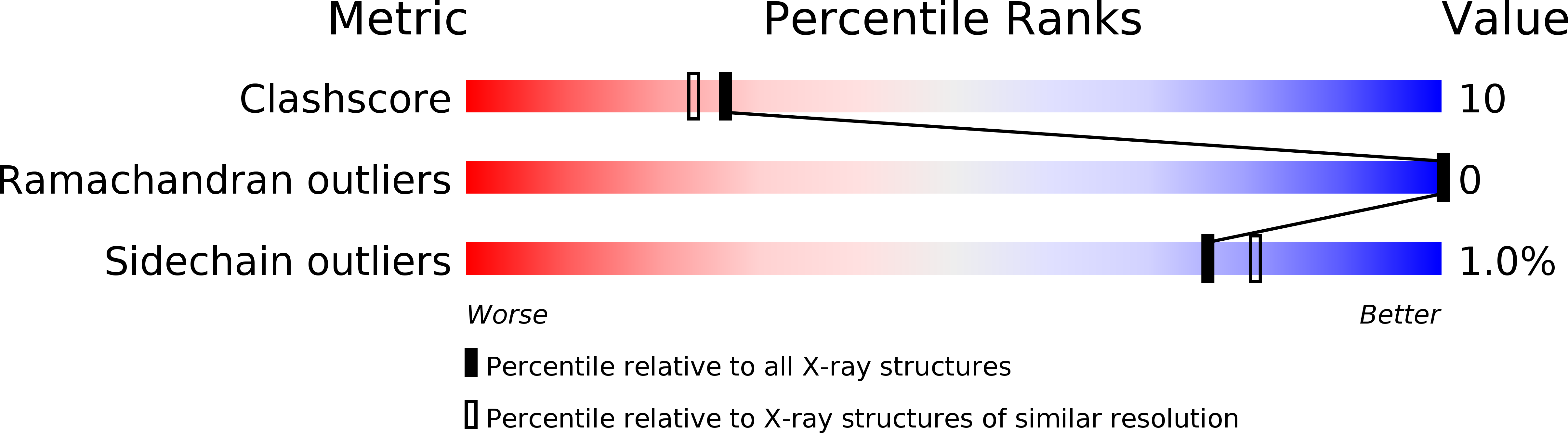Structure of the light chain-binding domain of myosin V.
Terrak, M., Rebowski, G., Lu, R.C., Grabarek, Z., Dominguez, R.(2005) Proc Natl Acad Sci U S A 102: 12718-12723
- PubMed: 16120677
- DOI: https://doi.org/10.1073/pnas.0503899102
- Primary Citation of Related Structures:
1N2D - PubMed Abstract:
Myosin V is a double-headed molecular motor involved in organelle transport. Two distinctive features of this motor, processivity and the ability to take extended linear steps of approximately 36 nm along the actin helical track, depend on its unusually long light chain-binding domain (LCBD). The LCBD of myosin V consists of six tandem IQ motifs, which constitute the binding sites for calmodulin (CaM) and CaM-like light chains. Here, we report the 2-A resolution crystal structure of myosin light chain 1 (Mlc1p) bound to the IQ2-IQ3 fragment of Myo2p, a myosin V from Saccharomyces cerevisiae. This structure, combined with FRET distance measurements between probes in various CaM-IQ complexes, comparative sequence analysis, and the previously determined structures of Mlc1p-IQ2 and Mlc1p-IQ4, allowed building a model of the LCBD of myosin V. The IQs of myosin V are distributed into three pairs. There appear to be specific cooperative interactions between light chains within each IQ pair, but little or no interaction between pairs, providing flexibility at their junctions. The second and third IQ pairs each present a light chain, whether CaM or a CaM-related molecule, bound in a noncanonical extended conformation in which the N-lobe does not interact with the IQ motif. The resulting free N-lobes may engage in protein-protein interactions. The extended conformation is characteristic of the single IQ of myosin VI and is common throughout the myosin superfamily. The model points to a prominent role of the LCBD in the function, regulation, and molecular interactions of myosin V.
Organizational Affiliation:
Boston Biomedical Research Institute, 64 Grove Street, Watertown, MA 02472, USA.