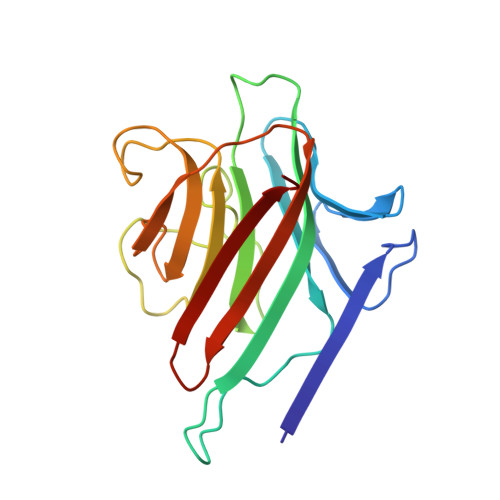
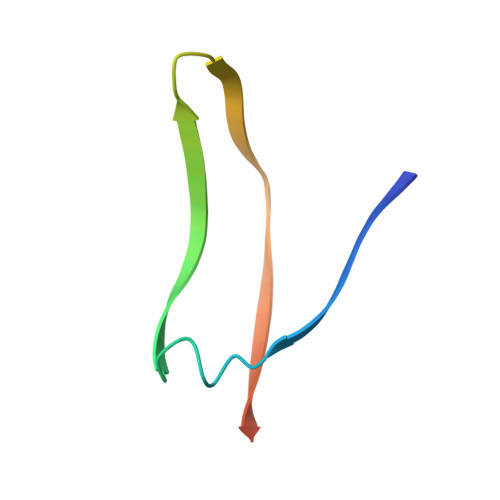
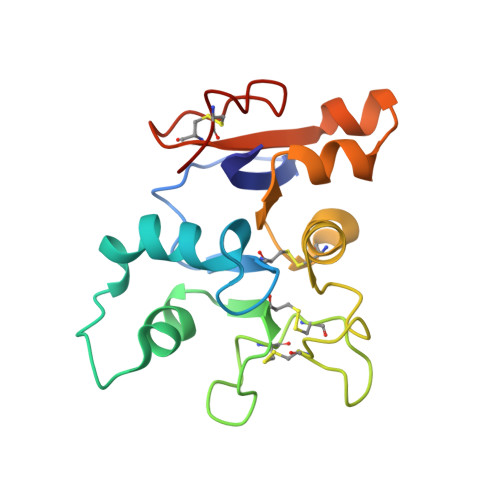
Structures of a legume lectin complexed with the human lactotransferrin N2 fragment, and with an isolated biantennary glycopeptide: role of the fucose moiety.
Bourne, Y., Mazurier, J., Legrand, D., Rouge, P., Montreuil, J., Spik, G., Cambillau, C.(1994) Structure 2: 209-219
- PubMed: 8069634
- DOI: https://doi.org/10.1016/s0969-2126(00)00022-8
- Primary Citation of Related Structures:
1LGB, 1LGC - PubMed Abstract:
Lectins mediate cell-cell interactions by specifically recognizing oligosaccharide chains. Legume lectins serve as mediators for the symbiotic interactions between plants and nitrogen-fixing microorganisms, an important process in the nitrogen cycle. Lectins from the Viciae tribe have a high affinity for the fucosylated biantennary N-acetyllactosamine-type glycans which are to be found in the majority of N-glycosylproteins. While the structures of several lectins complexed with incomplete oligosaccharides have been solved, no previous structure has included the complete glycoprotein. We have determined the crystal structures of Lathyrus ochrus isolectin II complexed with the N2 monoglycosylated fragment of human lactotransferrin (18 kDa) and an isolated glycopeptide (2.1 kDa) fragment of human lactotransferrin (at 3.3 A and 2.8 A resolution, respectively). Comparison between the two structures showed that the protein part of the glycoprotein has little influence on either the stabilization of the complex or the sugar conformation. In both cases the oligosaccharide adopts the same extended conformation. Besides the essential mannose moiety of the monosaccharide-binding site, the fucose-1' of the core has a large surface of interaction with the lectin. This oligosaccharide conformation differs substantially from that seen in the previously determined isolectin I-octasaccharide complex. Comparison of our structure with that of concanavalin A (ConA) suggests that the ConA binding site cannot accommodate this fucose. Our results explain the observation that Viciae lectins have a higher affinity for fucosylated oligosaccharides than for unfucosylated ones, whereas the affinity of ConA for these types of oligosaccharides is similar. This explanation is testable by mutagenesis experiments. Our structure shows a large complementary surface area between the oligosaccharide and the lectin, in contrast with the recently determined structure of a complex between the carbohydrate recognition domain of a C-type mammalian lectin and an oligomannoside, where only the non-reducing terminal mannose residue interacts with the lectin.
Organizational Affiliation:
Laboratoire de Cristallographie et de Cristallisation des Macromolécules Biologiques, CNRS URA 1296, Faculté de Médecine Secteur-Nord, Marseille, France.