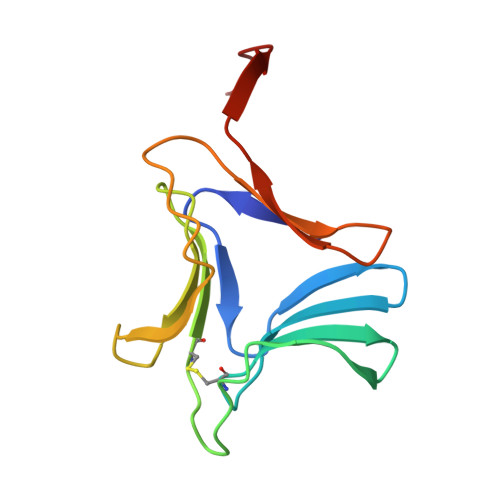
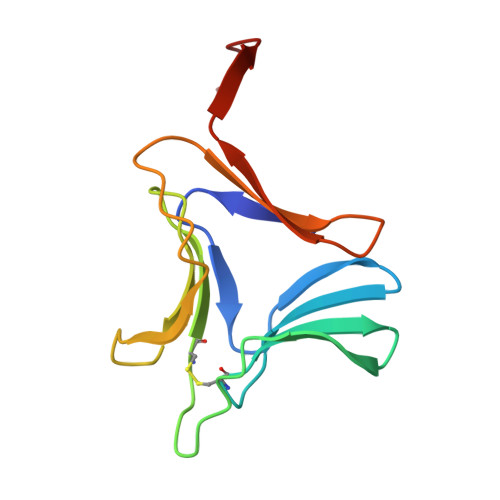
Re-refinement using reprocessed data to improve the quality of the structure: a case study involving garlic lectin.
Ramachandraiah, G., Chandra, N.R., Surolia, A., Vijayan, M.(2002) Acta Crystallogr D Biol Crystallogr 58: 414-420
- PubMed: 11856826
- DOI: https://doi.org/10.1107/s0907444901021497
- Primary Citation of Related Structures:
1KJ1 - PubMed Abstract:
The structure of dimeric garlic lectin was previously determined to an effective resolution of 2.8A using X-ray intensity data processed by the XDS package and refined using X-PLOR [Chandra et al. (1999), J. Mol. Biol. 285, 1157--1168]. Repeated attempts to grow better crystals with a view to improving the definition of the structure did not succeed. The available raw data were then reprocessed using DENZO. The structure was re-refined with both X-PLOR and CNS separately using the reprocessed data, which extended to a resolution of 2.2A. These two sets of refinements and the two sets using the XDS-processed data afforded an opportunity to compare the performance of different data-processing and refinement packages when dealing with data from weakly diffracting crystals. The best results were obtained when CNS was employed for refinement using data processed by DENZO. The quality and the resolution of the map and the definition of the structure improved substantially. In particular, the amino-acid residues at the variable locations in the sequence, and hence the isolectins, could be identified with a high degree of confidence. It could be established that the crystal asymmetric unit contains two identical heterodimers. The new refined structure also provided a better definition of other finer structural details.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560012, India.