A branched N-linked glycan at atomic resolution in the 1.12 A structure of rhamnogalacturonan acetylesterase.
Molgaard, A., Larsen, S.(2002) Acta Crystallogr D Biol Crystallogr 58: 111-119
- PubMed: 11752785
- DOI: https://doi.org/10.1107/s0907444901018479
- Primary Citation of Related Structures:
1K7C - PubMed Abstract:
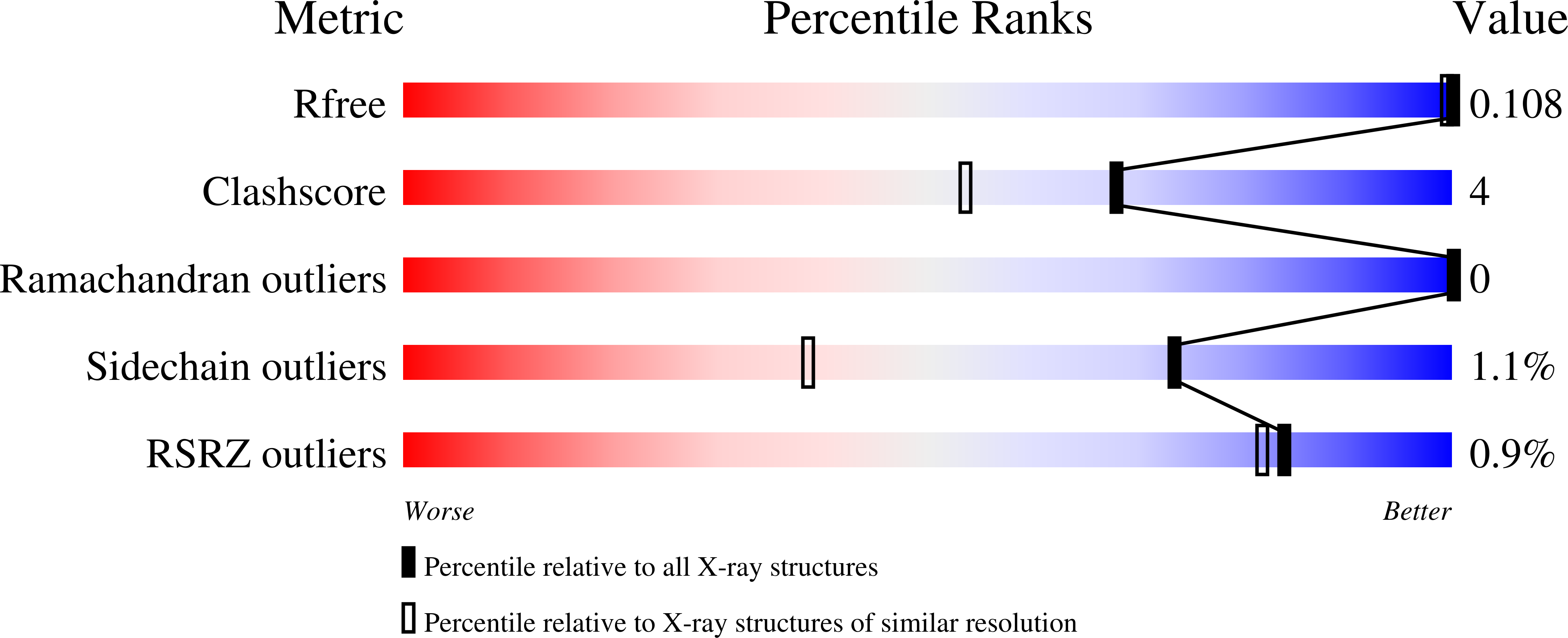
The crystal structure of the glycoprotein rhamnogalacturonan acetylesterase from Aspergillus aculeatus has been refined to a resolution of 1.12 A using synchrotron data collected at 263 K. Both of the two putative N-glycosylation sites at Asn104 and Asn182 are glycosylated and, owing to crystal contacts, the glycan structure at Asn182 is exceptionally well defined in the electron-density maps, showing the six-carbohydrate structure Manalpha1-6(Manalpha1-3)Manalpha1-6Manbeta1-4GlcNAcbeta1-4GlcNAcbeta-Asn182. Equivalent carbohydrate residues were restrained to have similar geometries, but were refined without target values. The refined bond lengths and angles were compared with the values obtained from small-molecule studies that form the basis for the dictionaries used for glycoprotein refinement.
Organizational Affiliation:
Centre for Crystallographic Studies, University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen, Denmark.