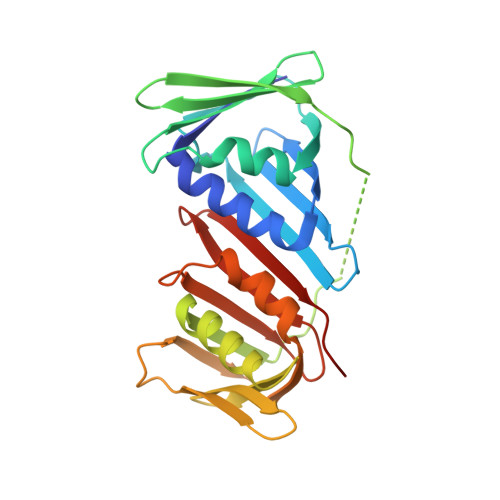
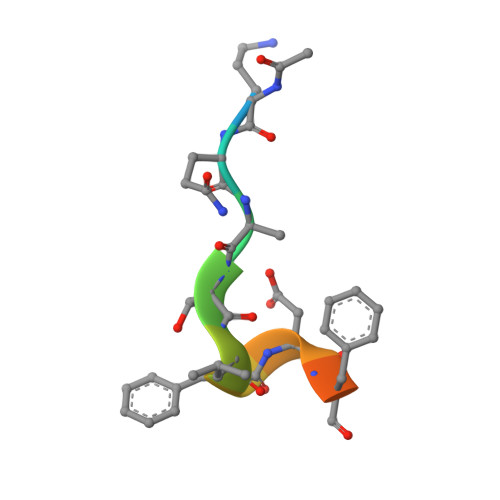
Physical interaction between proliferating cell nuclear antigen and replication factor C from Pyrococcus furiosus
Matsumiya, S., Ishino, S., Ishino, Y., Morikawa, K.(2002) Genes Cells 7: 911-922
- PubMed: 12296822
- DOI: https://doi.org/10.1046/j.1365-2443.2002.00572.x
- Primary Citation of Related Structures:
1ISQ - PubMed Abstract:
Proliferating cell nuclear antigen (PCNA), which is recognized as a DNA polymerase processivity factor, has direct interactions with various proteins involved in the important genetic information processes in Eukarya. We determined the crystal structure of PCNA from the hyperthermophilic archaeon, Pyrococcus furiosus (PfuPCNA) at 2.1 A resolution, and found that the toroidal ring-shaped structure, which consists of homotrimeric molecules, is highly conserved between the Eukarya and Archaea. This allowed us to examine its interaction with the loading factor at the atomic level. The replication factor C (RFC) is known as the loading factor of PCNA on to the DNA strand. P. furiosus RFC (PfuRFC) has a PCNA binding motif (PIP-box) at the C-terminus of the large subunit (RFCL). An 11 residue-peptide containing a PIP-box sequence of RFCL inhibited the PCNA-dependent primer extension ability of P. furiosus PolI in a concentration-dependent manner. To understand the molecular interaction mechanism of PCNA with PCNA binding proteins, we solved the crystal structure of PfuPCNA complexed with the PIP-box peptide. The interaction mode of the two molecules is remarkably similar to that of human PCNA and a peptide containing the PIP-box of p21(WAF1/CIP1). Moreover, the PIP-box binding may have some effect on the stability of the ring structure of PfuPCNA by some domain shift. Our structural analysis on PfuPCNA suggests that the interaction mode of the PIP-box with PCNA is generally conserved among the PCNA interacting proteins and that the functional meaning of the interaction via the PIP-box possibly depends on each protein. A movement of the C-terminal region of the PCNA monomer by PIP-box binding may cause the PCNA ring to be more rigid, suitable for its functions.
Organizational Affiliation:
Department of Structural Biology, Biomolecular Engineering Research Institute, 6-2-3, Furuedai, Suita, Osaka 565-0874, Japan.