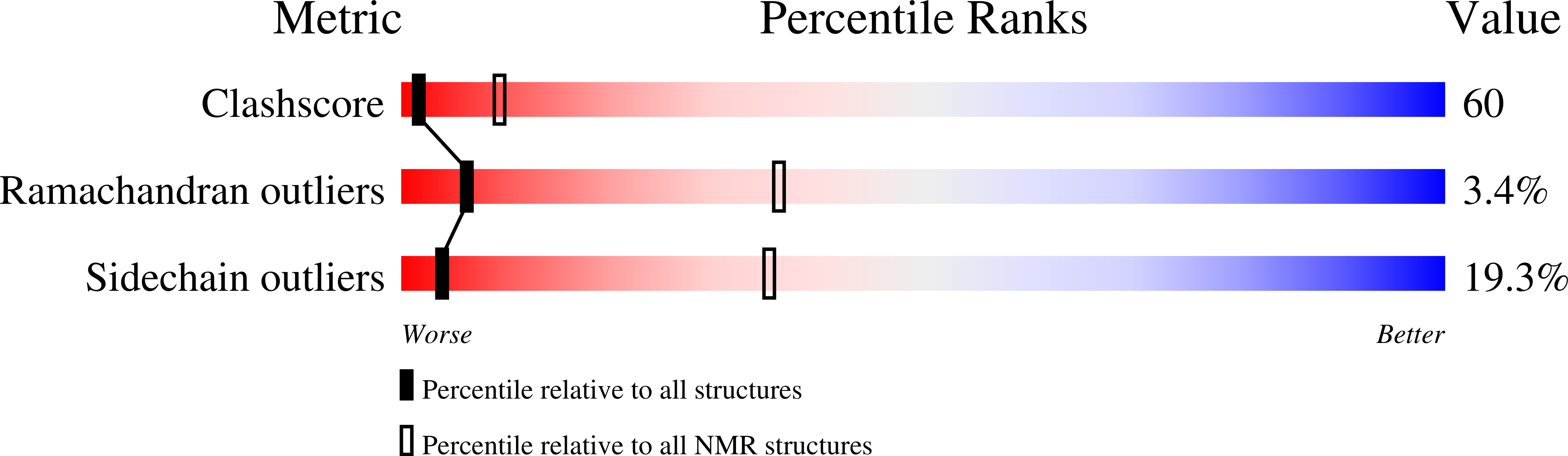High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Feher, V.A., Zapf, J.W., Hoch, J.A., Whiteley, J.M., McIntosh, L.P., Rance, M., Skelton, N.J., Dahlquist, F.W., Cavanagh, J.(1997) Biochemistry 36: 10015-10025
- PubMed: 9254596
- DOI: https://doi.org/10.1021/bi970816l
- Primary Citation of Related Structures:
1FSP, 2FSP - PubMed Abstract:
NMR has been employed for structural and dynamic studies of the bacterial response regulator, Spo0F. This 124-residue protein is an essential component of the sporulation phosphorelay signal transduction pathway in Bacillus subtilis. Three-dimensional 1H, 15N, and 13C experiments have been used to obtain full side chain assignments and the 1511 distance, 121 dihedral angle, and 80 hydrogen bonding restraints required for generating a family of structures (14 restraints per residue). The structures give a well-defined (alpha/beta)5 fold for residues 4-120 with average rms deviations of 0.59 A for backbone heavy atoms and 1.02 A for all heavy atoms. Analyses of backbone 15N relaxation measurements demonstrate relative rigidity in most regions of regular secondary structure with a generalized order parameter (S2) of 0.9 +/- 0.05 and a rotational correlation time (taum) of 7.0 +/- 0.5 ns. Loop regions near the site of phosphorylation have higher than average rms deviation values and T1/T2 ratios suggesting significant internal motion or chemical exchange at these sites. Additionally, multiple conformers are observed for the beta4-alpha4 loop and beta-strand 5 region. These conformers may be related to structural changes associated with phosphorylation and also indicative of the propensity this recognition surface has for differential protein interactions. Comparison of Spo0F structural features to those of other response regulators reveals subtle differences in the orientations of secondary structure in the putative recognition surfaces and the relative charge distribution of residues surrounding the site of phosphorylation. These may be important in providing specificity for protein-protein interactions and for determining the lifetimes of the phosphorylated state.
Organizational Affiliation:
Institute of Molecular Biology and Department of Chemistry, University of Oregon, Eugene, Oregon 97403, USA.