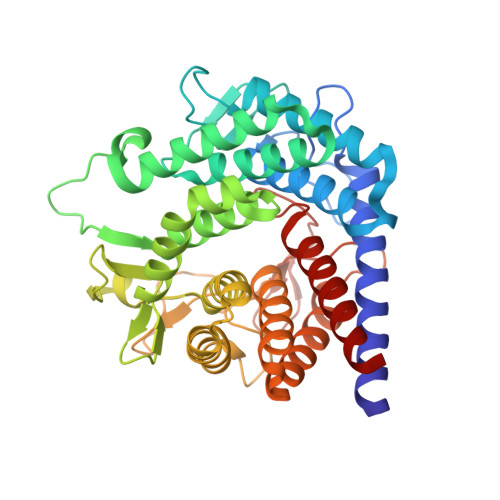
Crystal structure of N-acyl-D-glucosamine 2-epimerase from porcine kidney at 2.0 A resolution.
Itoh, T., Mikami, B., Maru, I., Ohta, Y., Hashimoto, W., Murata, K.(2000) J Mol Biol 303: 733-744
- PubMed: 11061972
- DOI: https://doi.org/10.1006/jmbi.2000.4188
- Primary Citation of Related Structures:
1FP3 - PubMed Abstract:
The X-ray crystallographic structure of N-acyl-d-glucosamine 2-epimerase (AGE) from porcine kidney, which has been identified to be a renin-binding protein (RnBP), was determined by the multiple isomorphous replacement method and refined at 2.0 A resolution with a final R-factor of 16.9 % for 15 to 2.0 A resolution data. The refined structure of AGE comprised 804 amino acid residues (one dimer) and 145 water molecules. The dimer of AGE had an asymmetric unit with approximate dimensions 46 Ax48 Ax96 A. The AGE monomer is composed of an alpha(6)/alpha(6)-barrel, the structure of which is found in glucoamylase and cellulase. One side of the AGE alpha(6)/alpha(6)-barrel structure comprises long loops containing five short beta-sheets, and contributes to the formation of a deep cleft shaped like a funnel. The putative active-site pocket and a possible binding site for the substrate N-acetyl-d-glucosamine (GlcNAc) were found in the cleft. The other side of the alpha(6)/alpha(6)-barrel comprises short loops and contributes to the dimer formation. At the dimer interface, which is composed of the short loops and alpha-helices of the subunits, five strong ion-pair interactions were observed, which play a major role in the dimer assembly. This completely ruled out the previously accepted hypothesis that the formation of the RnBP homodimer and RnBP-renin heterodimer requires the leucine zipper motif present in RnBP.
Organizational Affiliation:
Research Institute for Food Science, Uji Kyoto, 611-0011, Japan.