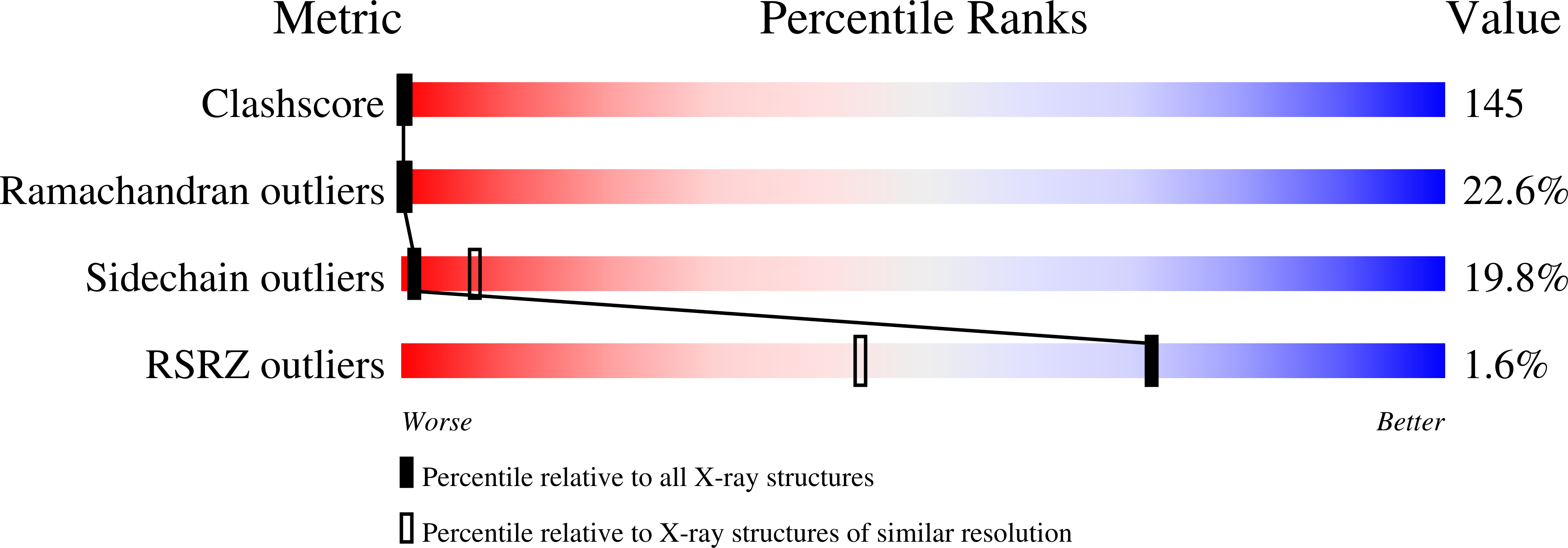
Recombinant cytochrome rC557 obtained from Escherichia coli cells expressing a truncated Thermus thermophilus cycA gene. Heme inversion in an improperly matured protein
McRee, D.E., Williams, P.A., Sridhar, V., Pastuszyn, A., Bren, K.L., Chen, Y., Patel, K.M., Todaro, T.R., Sanders, D., Luna, E., Fee, J.A.(2001) J Biol Chem 276: 6537-6544
- PubMed: 11069913
- DOI: https://doi.org/10.1074/jbc.M008421200
- Primary Citation of Related Structures:
1FOC - PubMed Abstract:
Cytochrome rC(557) is an improperly matured, dimeric cytochrome c obtained from expression of the "signal peptide-lacking" Thermus thermophilus cycA gene in the cytoplasm of Escherichia coli. It is characterized by its Q(00) (or alpha-) optical absorption band at 557 nm in the reduced form (Keightley, J. A., Sanders, D., Todaro, T. R., Pastuszyn, A., and Fee, J. A. (1998) J. Biol. Chem. 273, 12006-12016). We report results of a broad ranging, biochemical and spectral characterization of this protein that reveals the presence of a free vinyl group on the porphyrin and a disulfide bond between the protomers and supports His-Met ligation in both valence states of the iron. A 3-A resolution x-ray structure shows that, in comparison with the native protein, the heme moiety is rotated 180 degrees about its alpha,gamma-axis; cysteine 14 has formed a thioether bond with the 2-vinyl of pyrrole ring I instead of the 4-vinyl of pyrrole ring II, as occurs in the native protein; and a cysteine 11 from each protomer has formed an intermolecular disulfide bond. Numerous, minor perturbations exist within the structure of rC(557) in comparison with that of native protein, which result from heme inversion and protein-protein interactions across the dimer interface. The unusual spectral properties of rC(557) are rationalized in terms of this structure.
Organizational Affiliation:
Department of Molecular Biology, the Scripps Research Institute, La Jolla, California 92037, USA.