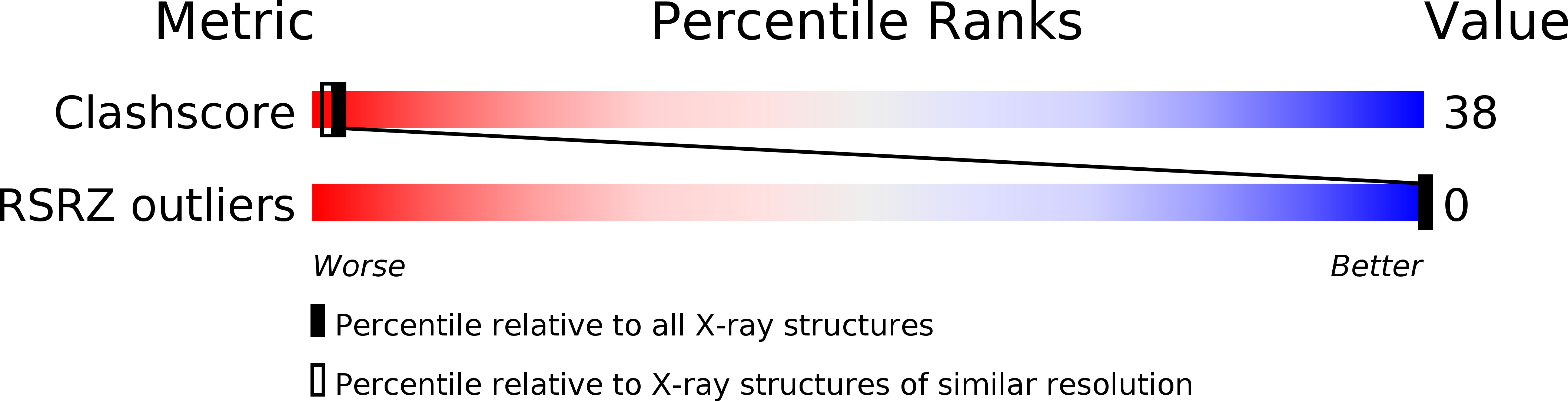Double helix conformation, groove dimensions and ligand binding potential of a G/C stretch in B-DNA.
Heinemann, U., Alings, C., Bansal, M.(1992) EMBO J 11: 1931-1939
- PubMed: 1582421
- DOI: https://doi.org/10.1002/j.1460-2075.1992.tb05246.x
- Primary Citation of Related Structures:
1CGC - PubMed Abstract:
The self-complementary DNA fragment CCGGCGCCGG crystallizes in the rhombohedral space group R3 with unit cell parameters a = 54.07 A and c = 44.59 A. The structure has been determined by X-ray diffraction methods at 2.2 A resolution and refined to an R value of 16.7%. In the crystal, the decamer forms B-DNA double helices with characteristic groove dimensions: compared with B-DNA of random sequence, the minor groove is wide and deep and the major groove is rather shallow. Local base pair geometries and stacking patterns are within the range commonly observed in B-DNA crystal structures. The duplex bears no resemblance to A-form DNA as might have been expected for a sequence with only GC base pairs. The shallow major groove permits an unusual crystal packing pattern with several direct intermolecular hydrogen bonds between phosphate oxygens and cytosine amino groups. In addition, decameric duplexes form quasi-infinite double helices in the crystal by end-to-end stacking. The groove geometries and accessibilities of this molecule as observed in the crystal may be important for the mode of binding of both proteins and drug molecules to G/C stretches in DNA.
Organizational Affiliation:
Institut für Kristallographie, Freie Universität Berlin, FRG.