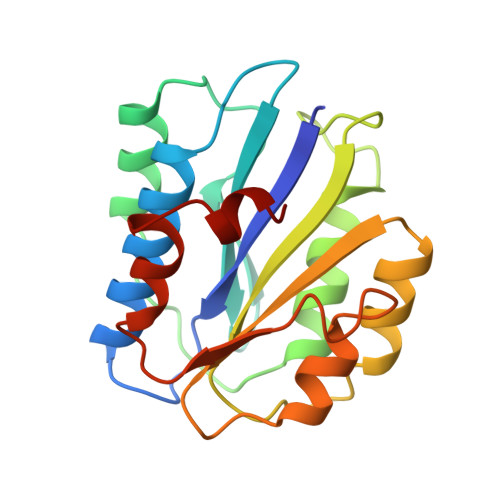
Crystal structure of the A3 domain of human von Willebrand factor: implications for collagen binding.
Huizinga, E.G., Martijn van der Plas, R., Kroon, J., Sixma, J.J., Gros, P.(1997) Structure 5: 1147-1156
- PubMed: 9331419
- DOI: https://doi.org/10.1016/s0969-2126(97)00266-9
- Primary Citation of Related Structures:
1ATZ - PubMed Abstract:
Bleeding from a damaged blood vessel is stopped by the formation of a platelet plug. The multimeric plasma glycoprotein, von Willebrand factor (vWF), plays an essential role in this process by anchoring blood platelets to the damaged vessel wall under conditions of high shear stress. This factor mediates platelet adhesion by binding both to collagen of the damaged blood vessel and to glycoprotein Ib on the platelet membrane. The A3 domain of vWF allows it to bind to collagen types I and III present in the perivascular connective tissue of the damaged vessel wall. To gain insight into the mechanism of collagen binding by vWF, we have determined the crystal structure of the human vWF A3 domain. The crystal structure of the 20 kDa A3 domain of human vWF (residues 920-1111), determined by the method of multiwavelength anomalous dispersion at 1.8 A resolution, exhibits a common dinucleotide-binding fold. The putative collagen-binding site of the A3 domain is rather smooth and shows a markedly high concentration of negatively charged residues. This region encompasses a potential metal-binding site containing the motif DXSXS, which is required for ligand interaction in the homologous I-type domains of integrins CR3 and LFA-1. Although vWF A3 has considerable sequence and structural similarity with CR3 and LFA-1 in this region, one loop of A3 adopts a conformation which is incompatible with ion binding. The structure of the A3 domain suggests that adhesion to collagen is primarily achieved through interactions between negatively charged residues on A3 and positively charged residues on collagen. The absence of a pronounced binding groove precludes a large van der Waals surface interaction between A3 and collagen and is consistent with the low affinity for collagen of a single A3 domain and the requirement for multimeric vWF for tight association with collagen. The absence of bound metal ions upon soaking the crystal in MgCl2 and vWF A3's conformational incompatibility for metal binding is consistent with the absence of a functional role for metal ion binding in A3, which contrasts the metal ion activation required for ligand binding by the homologous integrin I type domains.
Organizational Affiliation:
Department of Haematology, University Hospital Utrecht, The Netherlands.