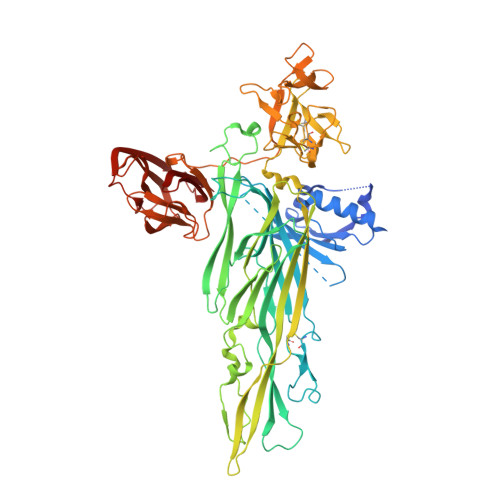
Crystal Structure of the Vibrio cholerae Cytolysin (VCC) Pro-toxin and its Assembly into a Heptameric Transmembrane Pore
Olson, R., Gouaux, E.(2005) J Mol Biol 350: 997-1016
- PubMed: 15978620
- DOI: https://doi.org/10.1016/j.jmb.2005.05.045
- Primary Citation of Related Structures:
1XEZ - PubMed Abstract:
Pathogenic Vibrio cholerae secrete V. cholerae cytolysin (VCC), an 80 kDa pro-toxin that assembles into an oligomeric pore on target cell membranes following proteolytic cleavage and interaction with cell surface receptors. To gain insight into the activation and targeting activities of VCC, we solved the crystal structure of the pro-toxin at 2.3A by X-ray diffraction. The core cytolytic domain of VCC shares a fold similar to the staphylococcal pore-forming toxins, but in VCC an amino-terminal pro-domain and two carboxy-terminal lectin domains decorate the cytolytic domain. The pro-domain masks a protomer surface that likely participates in inter-protomer interactions in the cytolytic oligomer, thereby explaining why proteolytic cleavage and movement of the pro-domain is necessary for toxin activation. A single beta-octyl glucoside molecule outlines a possible receptor binding site on one lectin domain, and removal of this domain leads to a tenfold decrease in lytic activity toward rabbit erythrocytes. VCC activated by proteolytic cleavage assembles into an oligomeric species upon addition of soybean asolectin/cholesterol liposomes and this oligomer was purified in detergent micelles. Analytical ultracentrifugation and crystallographic analysis indicate that the resulting VCC oligomer is a heptamer. Taken together, these studies define the architecture of a pore forming toxin and associated lectin domains, confirm the stoichiometry of the assembled oligomer as heptameric, and suggest a common mechanism of assembly for staphylococcal and Vibrio cytolytic toxins.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, 650 W. 168th Street, New York, NY 10032, USA.