Crystal structures of the GluR5 and GluR6 ligand binding cores: Molecular mechanisms underlying kainate receptor selectivity
Mayer, M.L.(2005) Neuron 45: 539-552
- PubMed: 15721240
- DOI: https://doi.org/10.1016/j.neuron.2005.01.031
- Primary Citation of Related Structures:
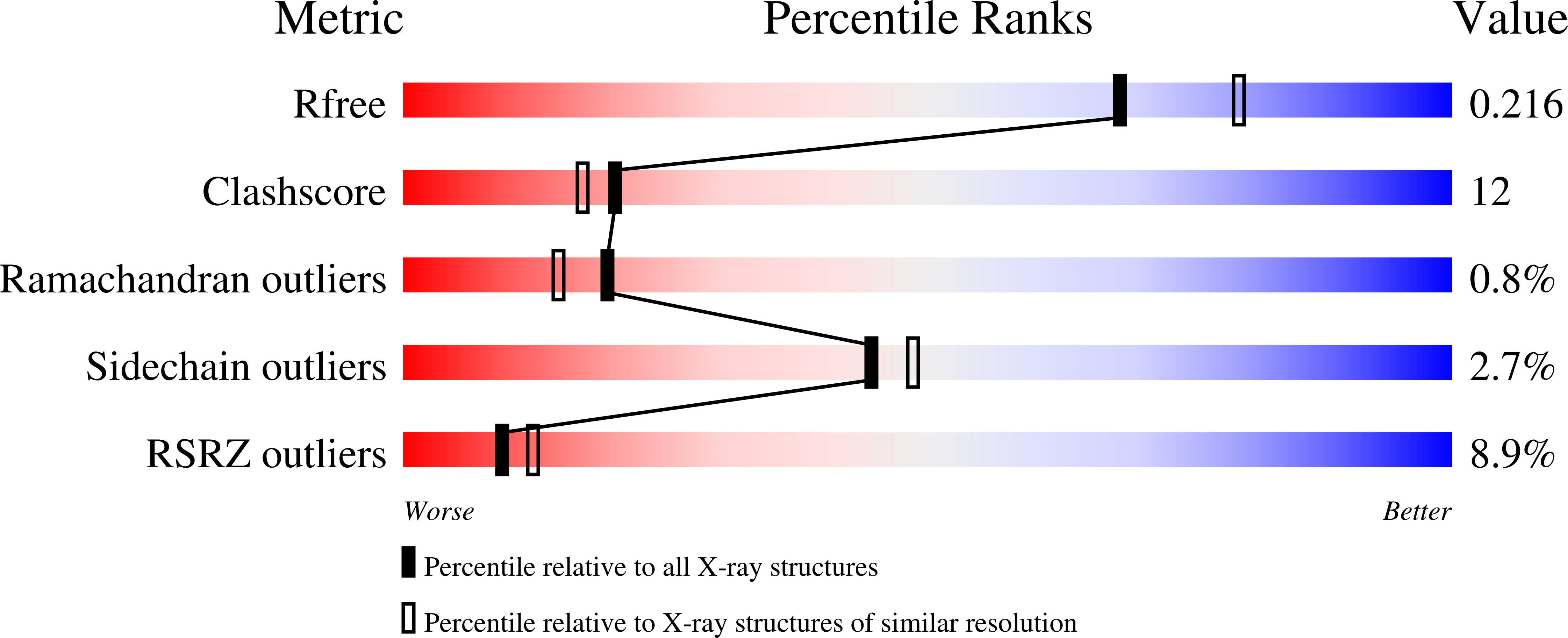
1S50, 1S7Y, 1S9T, 1SD3, 1TT1, 1TXF - PubMed Abstract:
Little is known about the molecular mechanisms underlying differences in the ligand binding properties of AMPA, kainate, and NMDA subtype glutamate receptors. Crystal structures of the GluR5 and GluR6 kainate receptor ligand binding cores in complexes with glutamate, 2S,4R-4-methylglutamate, kainate, and quisqualate have now been solved. The structures reveal that the ligand binding cavities are 40% (GluR5) and 16% (GluR6) larger than for GluR2. The binding of AMPA- and GluR5-selective agonists to GluR6 is prevented by steric occlusion, which also interferes with the high-affinity binding of 2S,4R-4-methylglutamate to AMPA receptors. Strikingly, the extent of domain closure produced by the GluR6 partial agonist kainate is only 3 degrees less than for glutamate and 11 degrees greater than for the GluR2 kainate complex. This, together with extensive interdomain contacts between domains 1 and 2 of GluR5 and GluR6, absent from AMPA receptors, likely contributes to the high stability of GluR5 and GluR6 kainate complexes.
Organizational Affiliation:
Porter Neuroscience Research Center, National Institute of Child Health and Human Development, National Institutes of Health, Department of Health and Human Services, Bethesda, Maryland 20892, USA. mlm@helix.nih.gov