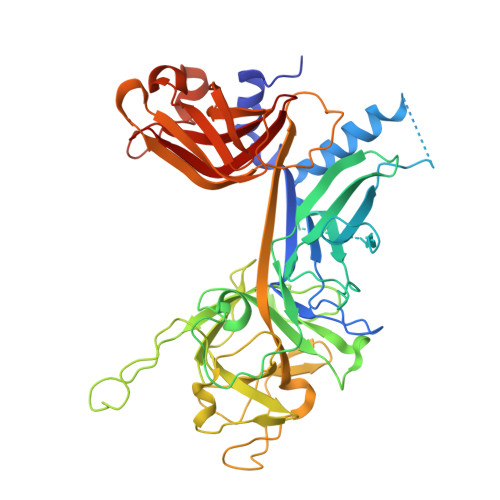
Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA
Kovall, R.A., Hendrickson, W.A.(2004) EMBO J 23: 3441-3451
- PubMed: 15297877
- DOI: https://doi.org/10.1038/sj.emboj.7600349
- Primary Citation of Related Structures:
1TTU - PubMed Abstract:
Notch signaling is a conserved pathway of communication between neighboring cells that results in cell fate specification, and CSL is the universal transcriptional effector of Notch signaling. The Notch intracellular domain translocates to the nucleus after proteolytic release upon Notch extracellular engagement, and there it displaces corepressors from DNA-bound CSL and recruits activators of Notch target genes. Here we report the 2.85 A crystal structure of CSL with a target DNA. CSL comprises three structurally integrated domains: its amino (NTD)- and carboxy (CTD)-terminal domains are strikingly similar to those of Rel transcription factors, but a surprising beta-trefoil domain (BTD) is inserted between them. CSL-bound DNA is recognized specifically by conserved residues from NTD and BTD. A hydrophobic pocket on BTD is identified as the likely site of Notch interaction with CSL, which has functional implications for the mechanism of Notch signaling.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Howard Hughes Medical Institute, Columbia University, New York, NY 10032, USA.