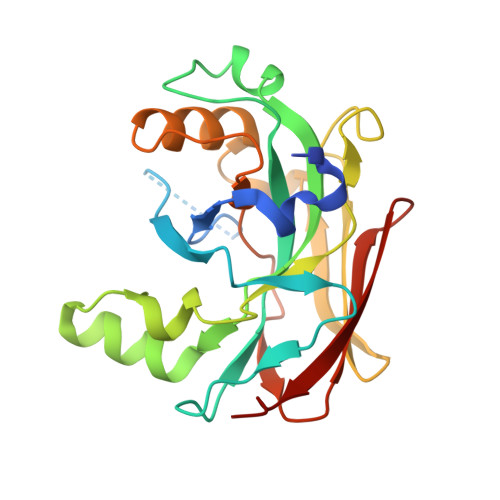
X-ray structure of fumarylacetoacetate hydrolase family member Homo sapiens FLJ36880.
Manjasetty, B.A., Niesen, F.H., Delbruck, H., Gotz, F., Sievert, V., Bussow, K., Behlke, J., Heinemann, U.(2004) Biol Chem 385: 935-942
- PubMed: 15551868
- DOI: https://doi.org/10.1515/BC.2004.122
- Primary Citation of Related Structures:
1SAW - PubMed Abstract:
The human protein FLJ36880 belongs to the fumarylacetoacetate hydrolase family. The X-ray structure of FLJ36880 has been determined to 2.2 A resolution employing the semi-automated high-throughput structural genomics approach of the Protein Structure Factory. FLJ36880 adopts a mixed beta-sandwich roll fold and forms homodimers in crystals as well as in solution. One Mg2+ ion is bound to each subunit of the dimeric protein by coordination to three carboxylate oxygens and three water molecules. These metal binding sites are accessible from the same surface of the dimer, partly due to the disorder of the undecapeptide stretch D29 to L39. The overall structure and metal binding site of FLJ36880 bear clear similarities to the C-terminal domain of the bifunctional enzyme HpcE from Escherichia coli C, fumarylacetoacetate hydrolase from Mus musculus and to YcgM (Apc5008) from E. coli 1262. These similarities provide a framework for suggesting biochemical functions and evolutionary relationships of FLJ36880. It appears highly probable that the metal binding sites are involved in an enzymatic activity related to the catabolism of aromatic amino acids. Two point mutations in the active-site of FAH, responsible for the metabolic disease hereditary tyrosinemia type I (HTI) in humans, affect residues that are structurally conserved in FLJ36880 and located in the putative catalytic site.
Organizational Affiliation:
Protein Structure Factory, c/o BESSY GmbH, Albert-Einstein-Str. 15, D-12489 Berlin, Germany.