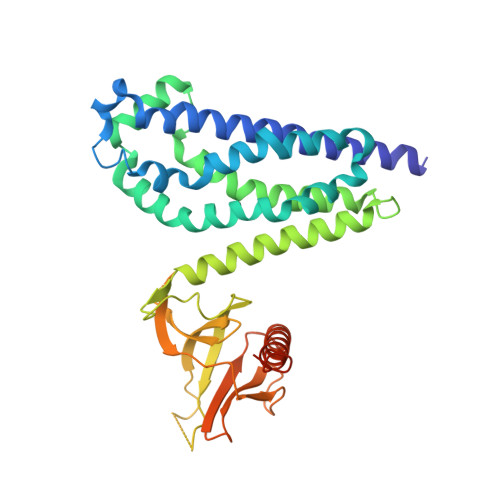
Crystal structure of the DH/PH fragment of Dbs without bound GTPase.
Worthylake, D.K., Rossman, K.L., Sondek, J.(2004) Structure 12: 1078-1086
- PubMed: 15274927
- DOI: https://doi.org/10.1016/j.str.2004.03.021
- Primary Citation of Related Structures:
1RJ2 - PubMed Abstract:
Dbl proteins are guanine nucleotide exchange factors for Rho GTPases, containing adjacent Dbl homology (DH) and pleckstrin homology (PH) domains. This domain architecture is virtually invariant and typically required for full exchange potential. Several structures of DH/PH fragments bound to GTPases implicate the PH domain in nucleotide exchange. To more fully understand the functional linkage between DH and PH domains, we have determined the crystal structure of the DH/PH fragment of Dbs without bound GTPase. This structure is generally similar to previously determined structures of Dbs bound to GTPases albeit with greater apparent mobility between the DH and PH domains. These comparisons suggest that the DH and PH domains of Dbs are spatially primed for binding GTPases and small alterations in intradomain conformations that may be elicited by subtle biological responses, such as altered phosphoinositide levels, are sufficient to enhance exchange by facilitating interactions between the PH domain and GTPases.
Organizational Affiliation:
Department of Pharmacology, University of North Carolina School of Medicine, Chapel Hill, NC 27599, USA.