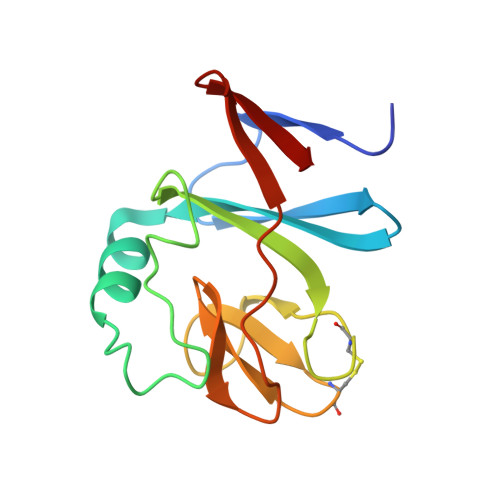
Structure of a water soluble fragment of the 'Rieske' iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 A resolution.
Iwata, S., Saynovits, M., Link, T.A., Michel, H.(1996) Structure 4: 567-579
- PubMed: 8736555
- DOI: https://doi.org/10.1016/s0969-2126(96)00062-7
- Primary Citation of Related Structures:
1RIE - PubMed Abstract:
The 'Rieske' iron-sulfur protein is the primary electron acceptor during hydroquinone oxidation in cytochrome bc complexes. The spectroscopic and electrochemical properties of the 'Rieske' [2Fe-2S] cluster differ significantly from those of other iron-sulfur clusters. A 129-residue water soluble fragment containing the intact [2Fe-2S] cluster was isolated following proteolytic digestion of the bc1 complex and used for structural studies. The structure of the Rieske iron-sulfur fragment containing the reduced [2Fe-2S] cluster has been determined using the multiwavelength anomalous diffraction (MAD) technique and refined at 1.5 A resolution. The fragment has a novel overall fold that includes three sheets of beta strands. The iron atoms of the [2Fe-2S] cluster are coordinated by two cysteine (Fe-1) and two histidine (Fe-2) residues, respectively, with the histidine ligands completely exposed to the solvent. This is in contrast to the four cysteine coordination pattern observed in previously characterised [2Fe-2S] ferredoxins. The cluster-binding fold is formed by two loops connected by a disulfide bridge; these loops superpose with the metal-binding loops of rubredoxins. The environment of the cluster is stabilised by an extensive hydrogen-bond network. The high-resolution structure supports the proposed coordination pattern involving histidine ligands and provides a basis for a detailed analysis of the spectroscopic and electrochemical properties. As the cluster is located at the tip of the protein, it might come into close contact with cytochrome b. The exposed N epsilon atoms of the histidine ligands of the cluster are readily accessible to quinones and inhibitors within the hydroquinone oxidation (QP) pocket of the bc1 complex and may undergo redox-dependent protonation/deprotonation.
Organizational Affiliation:
Max-Planck-Institut für Biophysik, Abt. Molekulare Membranbiologie, Frankfurt/Main., Germany.