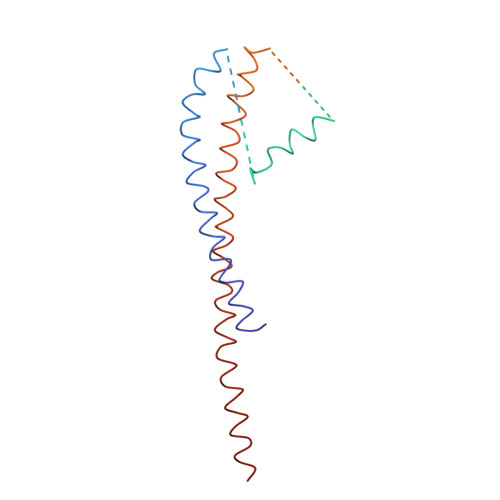Molecular Architecture of the Rotary Motor in ATP Synthase
Stock, D., Leslie, A.G.W., Walker, J.E.(1999) Science 286: 1700
- PubMed: 10576729
- DOI: https://doi.org/10.1126/science.286.5445.1700
- Primary Citation of Related Structures:
1QO1, 2XOK - PubMed Abstract:
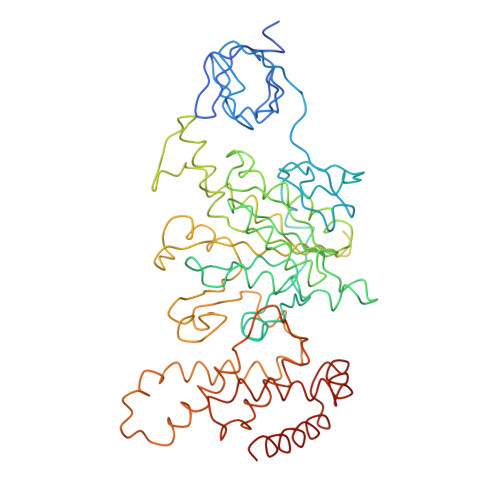
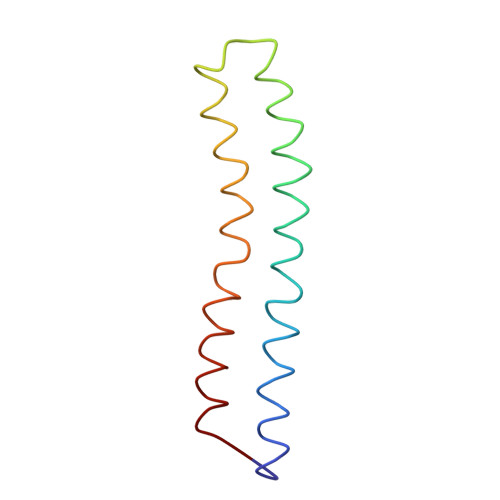
Adenosine triphosphate (ATP) synthase contains a rotary motor involved in biological energy conversion. Its membrane-embedded F0 sector has a rotation generator fueled by the proton-motive force, which provides the energy required for the synthesis of ATP by the F1 domain. An electron density map obtained from crystals of a subcomplex of yeast mitochondrial ATP synthase shows a ring of 10 c subunits. Each c subunit forms an alpha-helical hairpin. The interhelical loops of six to seven of the c subunits are in close contact with the gamma and delta subunits of the central stalk. The extensive contact between the c ring and the stalk suggests that they may rotate as an ensemble during catalysis.
Organizational Affiliation:
Medical Research Council Dunn Human Nutrition Unit, Hills Road, Cambridge CB2 2XY, UK.