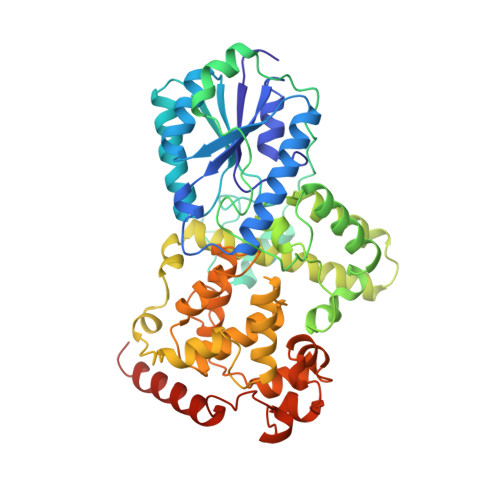
Crystal structure of DNA photolyase from Anacystis nidulans
Tamada, T., Kitadokoro, K., Higuchi, Y., Inaka, K., Yasui, A., de Ruiter, P.E., Eker, A.P., Miki, K.(1997) Nat Struct Biol 4: 887-891
- PubMed: 9360600
- DOI: https://doi.org/10.1038/nsb1197-887
- Primary Citation of Related Structures:
1QNF - PubMed Abstract:
The crystal structure at 1.8 A resolution of 8-HDF type photolyase from A. nidulans shows a backbone structure similar to that of MTHF type E. coli photolyase but reveals a completely different binding site for the light-harvesting cofactor.