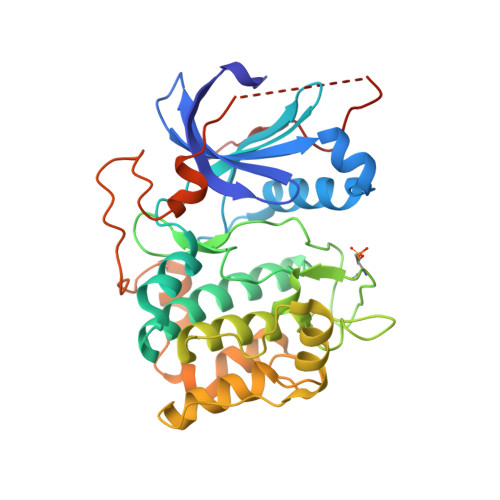
Crystal Structure of an Activated Akt/Protein Kinase B Ternary Complex with Gsk-3 Peptide and AMP-Pnp
Yang, J., Cron, P., Good, V.M., Thompson, V., Hemmings, B.A., Barford, D.(2002) Nat Struct Biol 9: 940
- PubMed: 12434148
- DOI: https://doi.org/10.1038/nsb870
- Primary Citation of Related Structures:
1O6K, 1O6L - PubMed Abstract:
The protein kinase Akt/PKB is stimulated by the phosphorylation of two regulatory residues, Thr 309 of the activation segment and Ser 474 of the hydrophobic motif (HM), that are structurally and functionally conserved within the AGC kinase family. To understand the mechanism of PKB regulation, we determined the crystal structures of activated kinase domains of PKB in complex with a GSK3beta-peptide substrate and an ATP analog. The activated state of the kinase was generated by phosphorylating Thr 309 using PDK1 and mimicking Ser 474 phosphorylation either with the S474D substitution or by replacing the HM of PKB with that of PIFtide, a potent mimic of a phosphorylated HM. Comparison with the inactive PKB structure indicates that the role of Ser 474 phosphorylation is to promote the engagement of the HM with the N-lobe of the kinase domain, promoting a disorder-to-order transition of the alphaC helix. The alphaC helix, by interacting with pThr 309, restructures and orders the activation segment, generating an active kinase conformation. Analysis of the interactions between PKB and the GSK3beta-peptide explains how PKB selects for protein substrates distinct from those of PKA.
Organizational Affiliation:
Section of Structural Biology, Institute of Cancer Research, Chester Beatty Laboratories, 237 Fulham Road, London, SW3 6JB, UK.