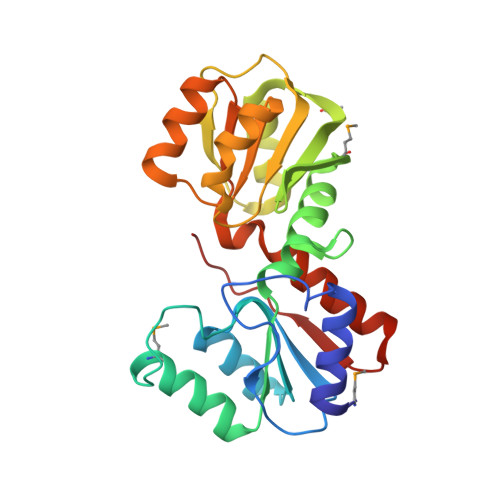
Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
Ladner, J.E., Obmolova, G., Teplyakov, A., Howard, A.J., Khil, P.P., Camerini-Otero, R.D., Gilliland, G.L.(2003) BMC Struct Biol 3: 7
- PubMed: 14519207
- DOI: https://doi.org/10.1186/1472-6807-3-7
- Primary Citation of Related Structures:
1LQA, 1NMO, 1NMP - PubMed Abstract:
The protein encoded by the gene ybgI was chosen as a target for a structural genomics project emphasizing the relation of protein structure to function.
Organizational Affiliation:
Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute and the National Institute of Standards and Technology, 9600 Gudelsky Drive, Rockville, MD 20850, USA. jane.ladner@nist.gov