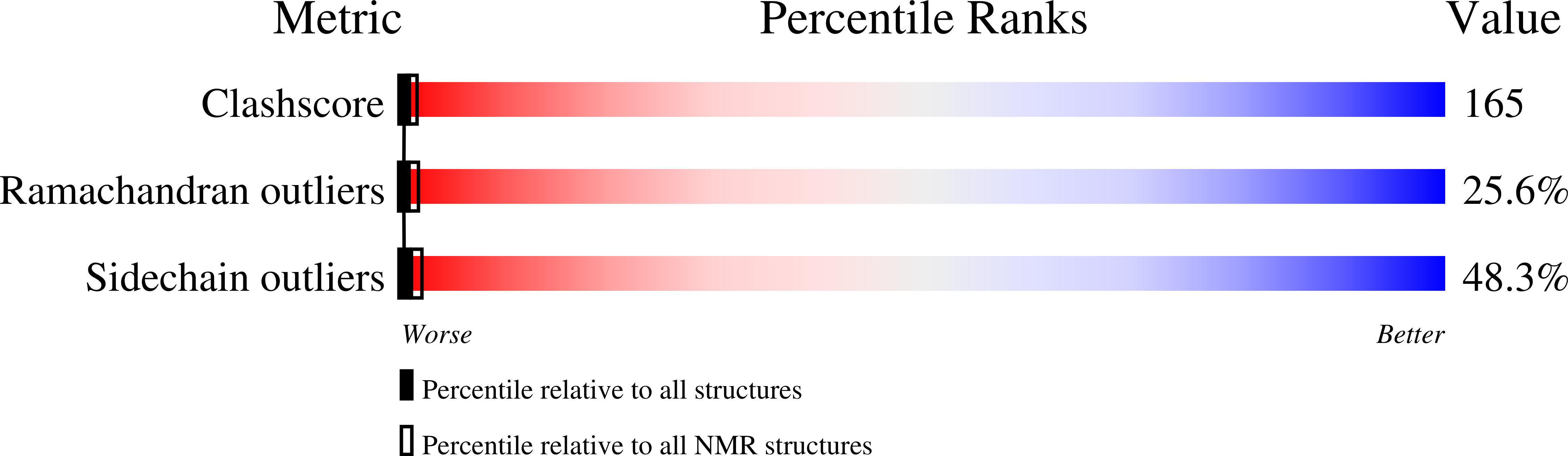Conformational preferences and activities of peptides from the catecholamine release-inhibitory (catestatin) region of chromogranin A
Preece, N.E., Nguyen, M., Mahata, M., Mahata, S.K., Mahapatra, N.R., Tsigelny, I., O'Connor, D.T.(2004) Regul Pept 118: 75-87
- PubMed: 14759560
- DOI: https://doi.org/10.1016/j.regpep.2003.10.035
- Primary Citation of Related Structures:
1LV4, 1N2Y - PubMed Abstract:
Previous modeling (PDB 1cfk) of the catecholamine release-inhibitory "catestatin" region of chromogranin A (CgA) suggested a beta-strand/loop/beta-strand active conformation, displaying an electropositive Arg-rich loop (R(351)AR(353)GYGFR(358)). To explore this possibility, we studied NMR structures of linear and cyclic synthetic catestatin, bovine (bCgA(344-364)) or human (hCgA(352-372)). By 2-D (1)H-NMR, the structure of linear catestatin (hCgA(352-372)) exhibited the NOE pattern of a coiled loop (PDB 1lv4). We then constrained the structure, cyclizing the putative Arg-rich loop connecting the beta-strands: cyclic bCgA(350-362) ([C(0)]F(350)RARGYGFRGPGL(362)[C(+14)]). Favored conformations of cyclic bCgA(350-362) were determined by (1)H-NMR and (13)C-NMR spectroscopy. Cyclic bCgA(350-362) conformers (PDB 1n2y) adopted a "twisted-loop" conformation. Alignment between the homology model and the cyclic NMR structure showed that, while portions of the NMR structure's mid-molecule and carboxy-terminus were congruent with the homology model (RMSD, 1.61-1.91 A), the amino-terminal "twisted loop" coiled inward and away from the model (RMSD, 3.36 A). Constrained cyclic bCgA(350-362) did not exert nicotinic cholinergic antagonist activity (IC(50)>10 microM), when compared to full-length linear (IC(50) approximately 0.42-0.56 microM), or cyclic (IC(50) approximately 0.74 microM) catestatin. Thus, loss of activity in the small, constrained peptide did not result from either [Cys]-extension or cyclization, per se. While linear catestatin displays coiled character, a small cyclic derivative lost biological activity, perhaps because its amino-terminal domain deviated sharply from the predicted active conformation. These results refine the relationship between structure and function in catestatin, and suggest goals in future peptidomimetic syntheses, in particular attempts to constrain the correct amino-terminal shape for biological activity.
Organizational Affiliation:
Department of Medicine, University of California, San Diego, and VA San Diego Healthcare System, 92161, USA.