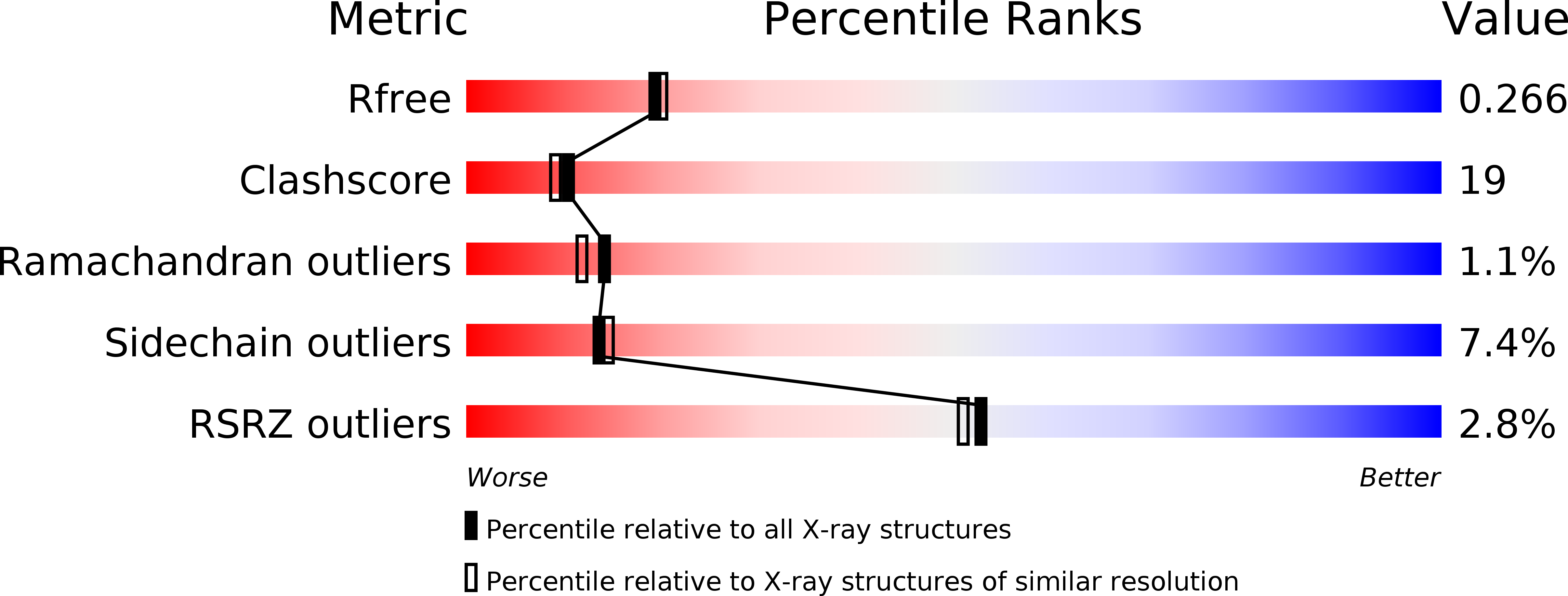
Crystal structure of GGA2 VHS domain and its implication in plasticity in the ligand binding pocket
Zhu, G., He, X., Terzyan, S., Zhai, P., Tang, J., Zhang, X.C.(2003) FEBS Lett 537: 171-176
- PubMed: 12606052
- DOI: https://doi.org/10.1016/s0014-5793(03)00095-4
- Primary Citation of Related Structures:
1MHQ - PubMed Abstract:
Golgi-localized, gamma-ear-containing, ARF binding (GGA) proteins regulate intracellular vesicle transport by recognizing sorting signals on the cargo surface in the initial step of the budding process. The VHS (VPS27, Hrs, and STAM) domain of GGA binds with the signal peptides. Here, a crystal structure of the VHS domain of GGA2 is reported at 2.2 A resolution, which permits a direct comparison with that of homologous proteins, GGA1 and GGA3. Significant structural difference is present in the loop between helices 6 and 7, which forms part of the ligand binding pocket. Intrinsic fluorescence spectroscopic study indicates that this loop undergoes a conformational change upon ligand binding. Thus, the current structure suggests that a conformational change induced by ligand binding occurs in this part of the ligand pocket.
Organizational Affiliation:
Crystallography Research Program of Oklahoma Medical Research Foundation, 825 N.E. 13th Street, Oklahoma City 73104, USA.