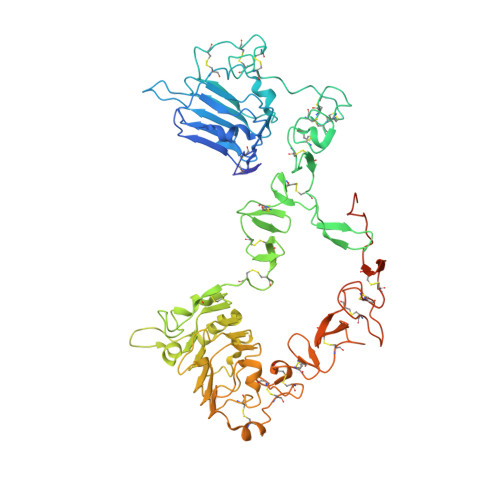
Structure of the extracellular region of HER3 reveals an interdomain tether.
Cho, H.S., Leahy, D.J.(2002) Science 297: 1330-1333
- PubMed: 12154198
- DOI: https://doi.org/10.1126/science.1074611
- Primary Citation of Related Structures:
1M6B - PubMed Abstract:
We have determined the 2.6 angstrom crystal structure of the entire extracellular region of human HER3 (ErbB3), a member of the epidermal growth factor receptor (EGFR) family. The structure consists of four domains with structural homology to domains found in the type I insulin-like growth factor receptor. The HER3 structure reveals a contact between domains II and IV that constrains the relative orientations of ligand-binding domains and provides a structural basis for understanding both multiple-affinity forms of EGFRs and conformational changes induced in the receptor by ligand binding during signaling. These results also suggest new therapeutic approaches to modulating the behavior of members of the EGFR family.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry, Howard Hughes Medical Institute, Johns Hopkins University School of Medicine, 725 North Wolfe Street, Baltimore, MD 21205, USA.