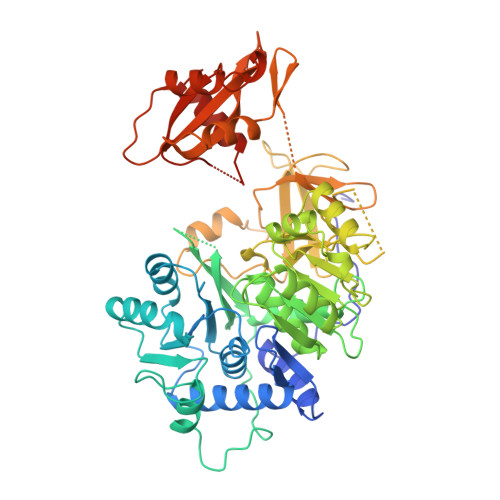
Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes.
Conti, E., Franks, N.P., Brick, P.(1996) Structure 4: 287-298
- PubMed: 8805533
- DOI: https://doi.org/10.1016/s0969-2126(96)00033-0
- Primary Citation of Related Structures:
1LCI - PubMed Abstract:
Firefly luciferase is a 62 kDa protein that catalyzes the production of light. In the presence of MgATP and molecular oxygen, the enzyme oxidizes its substrate, firefly luciferin, emitting yellow-green light. The reaction proceeds through activation of the substrate to form an adenylate intermediate. Firefly luciferase shows extensive sequence homology with a number of enzymes that utilize ATP in adenylation reactions. We have determined the crystal structure of firefly luciferase at 2.0 A resolution. The protein is folded into two compact domains. The large N-terminal domain consists of a beta-barrel and two beta-sheets. The sheets are flanked by alpha-helices to form an alphabetaalphabetaalpha five-layered structure. The C-terminal portion of the molecule forms a distinct domain, which is separated from the N-terminal domain by a wide cleft. Firefly luciferase is the first member of a superfamily of homologous enzymes, which includes acyl-coenzyme A ligases and peptide synthetases, to have its structure characterized. The residues conserved within the superfamily are located on the surfaces of the two domains on either side of the cleft, but are too far apart to interact simultaneously with the substrates. This suggests that the two domains will close in the course of the reaction. Firefly luciferase has a novel structural framework for catalyzing adenylate-forming reactions.
Organizational Affiliation:
Biophysics Section, Blackett Laboratory, Imperial College, London SW7 2BZ, UK.