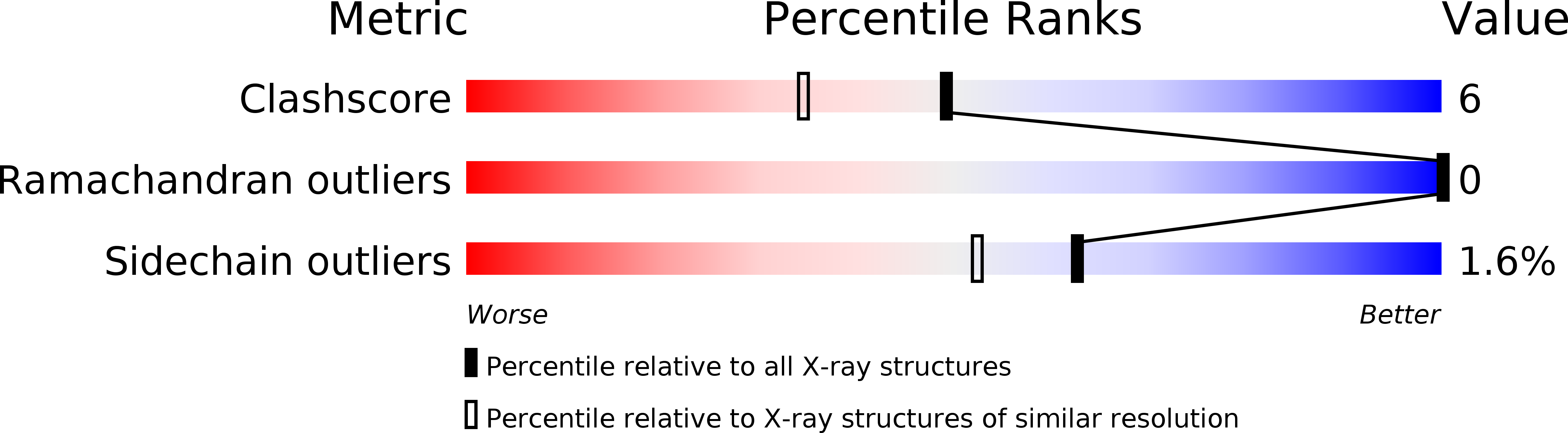Factorising ligand affinity: a combined thermodynamic and crystallographic study of trypsin and thrombin inhibition.
Dullweber, F., Stubbs, M.T., Musil, D., Sturzebecher, J., Klebe, G.(2001) J Mol Biol 313: 593-614
- PubMed: 11676542
- DOI: https://doi.org/10.1006/jmbi.2001.5062
- Primary Citation of Related Structures:
1K1I, 1K1J, 1K1L, 1K1M, 1K1N, 1K1O, 1K1P, 1K21, 1K22 - PubMed Abstract:
The binding of a series of low molecular weight ligands towards trypsin and thrombin has been studied by isothermal titration calorimetry and protein crystallography. In a series of congeneric ligands, surprising changes of protonation states occur and are overlaid on the binding process. They result from induced pK(a) shifts depending on the local environment experienced by the ligand and protein functional groups in the complex (induced dielectric fit). They involve additional heat effects that must be corrected before any conclusion on the binding enthalpy (DeltaH) and entropy (DeltaS) can be drawn. After correction, trends in both contributions can be interpreted in structural terms with respect to the hydrogen bond inventory or residual ligand motions. For all inhibitors studied, a strong negative heat capacity change (DeltaC(p)) is detected, thus binding becomes more exothermic and entropically less favourable with increasing temperature. Due to a mutual compensation, Gibbs free energy remains virtually unchanged. The strong negative DeltaC(p) value cannot solely be explained by the removal of hydrophobic surface portions of the protein or ligand from water exposure. Additional contributions must be considered, presumably arising from modulations of the local water structure, changes in vibrational modes or other ordering parameters. For thrombin, smaller negative DeltaC(p) values are observed for ligand binding in the presence of sodium ions compared to the other alkali ions, probably due to stabilising effects on the protein or changes in the bound water structure.
Organizational Affiliation:
Philipps-Universität Marburg, Institut für Pharmazeutische Chemie, Marbacher Weg 6, D-35037 Marburg (Lahn), Germany.