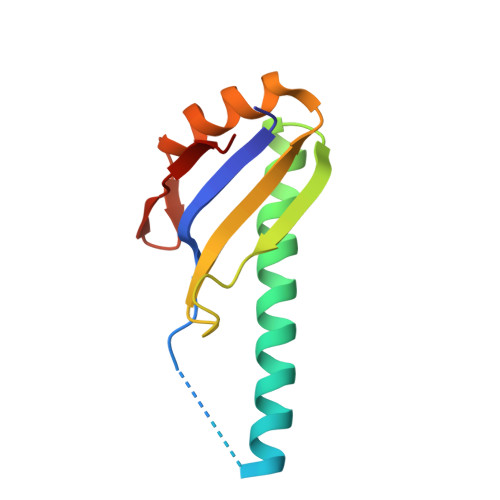
A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer.
Kielkopf, C.L., Rodionova, N.A., Green, M.R., Burley, S.K.(2001) Cell 106: 595-605
- PubMed: 11551507
- DOI: https://doi.org/10.1016/s0092-8674(01)00480-9
- Primary Citation of Related Structures:
1JMT - PubMed Abstract:
U2 auxiliary factor (U2AF) is an essential splicing factor that recognizes the 3' splice site and recruits the U2 snRNP to the branch point. The X-ray structure of the human core U2AF heterodimer, consisting of the U2AF35 central domain and a proline-rich region of U2AF65, has been determined at 2.2 A resolution. The structure reveals a novel protein-protein recognition strategy, in which an atypical RNA recognition motif (RRM) of U2AF35 and the U2AF65 polyproline segment interact via reciprocal "tongue-in-groove" tryptophan residues. Complementary biochemical experiments demonstrate that the core U2AF heterodimer binds RNA, and that the interacting tryptophan side chains are essential for U2AF dimerization. Atypical RRMs in other splicing factors may serve as protein-protein interaction motifs elsewhere during spliceosome assembly.
Organizational Affiliation:
Laboratories of Molecular Biophysics, The Rockefeller University, 1230 York Avenue, New York, NY 10021, USA.