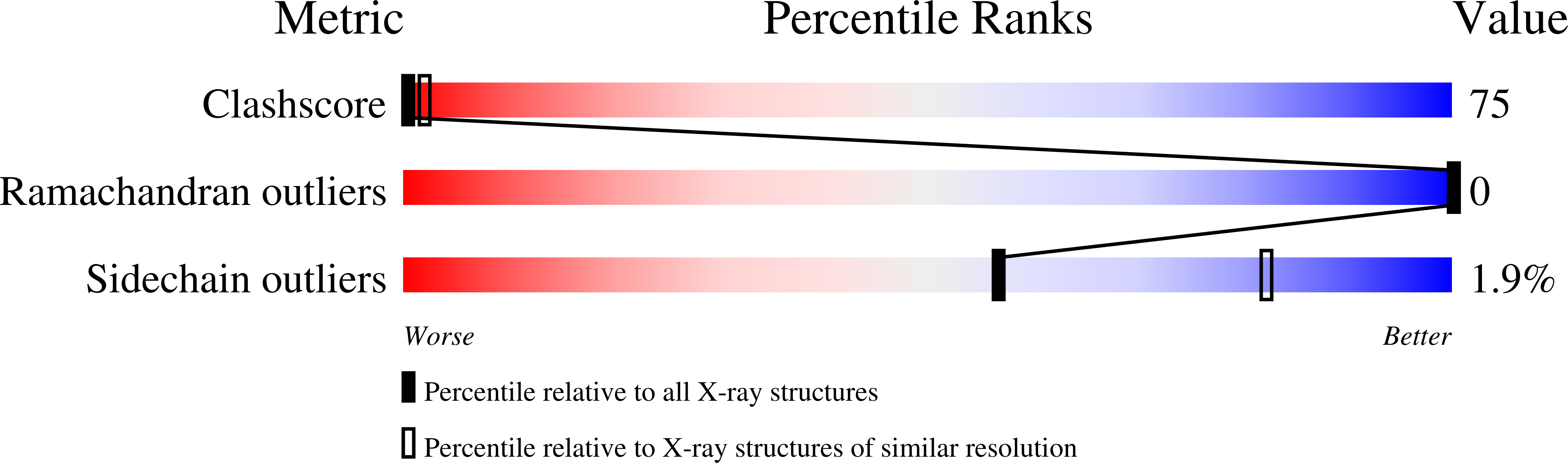Binding of N-acetylglucosamine to chicken egg lysozyme: a powder diffraction study.
Von Dreele, R.B.(2001) Acta Crystallogr D Biol Crystallogr 57: 1836-1842
- PubMed: 11717496
- DOI: https://doi.org/10.1107/s0907444901015748
- Primary Citation of Related Structures:
1JA2, 1JA4, 1JA6, 1JA7 - PubMed Abstract:
The binding of N-acetylglucosamine (NAG) to chicken egg lysozyme (E.C. 3.2.1.17) was investigated by high-resolution X-ray powder diffraction at room temperature. NAG was found to bind to lysozyme in a rapid precipitation preparation with 0.05 M NaCl buffer pH 6.0, but not 0.05 M NaCl buffer pH 5.0. Binding was indicated by significant and readily apparent changes in the diffraction pattern from that of the apo protein precipitated from the same solvent. The location of NAG bound to lysozyme was easily found from a difference Fourier map generated from structure factors extracted during a preliminary combined Rietveld and stereochemical restraint refinement. Full protein and protein-NAG structures were refined with these techniques (R(wp) = 2.22-2.49%, R(p) = 1.79-1.95%, R(F)(2) = 4.95-6.35%) and revealed a binding mode for NAG which differed from that found in an earlier single-crystal study and probably represents a precursor trapped by rapid precipitation.
Organizational Affiliation:
Manuel Lujan Jr Neutron Scattering Center, MS H805, Los Alamos National Laboratory, Los Alamos, NM 87545, USA. vondreele@lanl.gov