Mechanisms for ligand binding to GluR0 ion channels: crystal structures of the glutamate and serine complexes and a closed apo state.
Mayer, M.L., Olson, R., Gouaux, E.(2001) J Mol Biol 311: 815-836
- PubMed: 11518533
- DOI: https://doi.org/10.1006/jmbi.2001.4884
- Primary Citation of Related Structures:
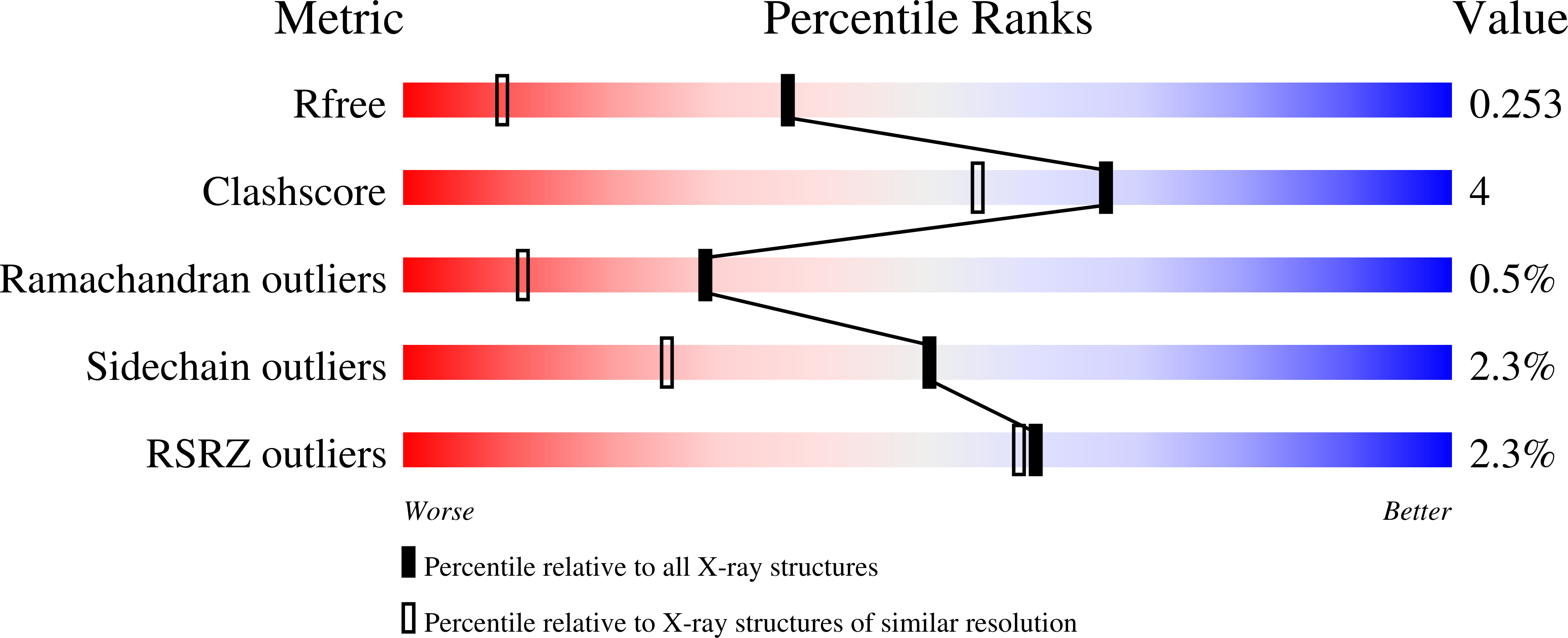
1II5, 1IIT, 1IIW - PubMed Abstract:
High-resolution structures of the ligand binding core of GluR0, a glutamate receptor ion channel from Synechocystis PCC 6803, have been solved by X-ray diffraction. The GluR0 structures reveal homology with bacterial periplasmic binding proteins and the rat GluR2 AMPA subtype neurotransmitter receptor. The ligand binding site is formed by a cleft between two globular alpha/beta domains. L-Glutamate binds in an extended conformation, similar to that observed for glutamine binding protein (GlnBP). However, the L-glutamate gamma-carboxyl group interacts exclusively with Asn51 in domain 1, different from the interactions of ligand with domain 2 residues observed for GluR2 and GlnBP. To address how neutral amino acids activate GluR0 gating we solved the structure of the binding site complex with L-serine. This revealed solvent molecules acting as surrogate ligand atoms, such that the serine OH group makes solvent-mediated hydrogen bonds with Asn51. The structure of a ligand-free, closed-cleft conformation revealed an extensive hydrogen bond network mediated by solvent molecules. Equilibrium centrifugation analysis revealed dimerization of the GluR0 ligand binding core with a dissociation constant of 0.8 microM. In the crystal, a symmetrical dimer involving residues in domain 1 occurs along a crystallographic 2-fold axis and suggests that tetrameric glutamate receptor ion channels are assembled from dimers of dimers. We propose that ligand-induced conformational changes cause the ion channel to open as a result of an increase in domain 2 separation relative to the dimer interface.
Organizational Affiliation:
Laboratory of Cellular and MolecularNeurophysiology, National Institute of Child Health and Human Development, National Institutes of Health, Building 49, Room 5A78, 49 Convent Drive MSC 4495, Bethesda, MD 20892, USA. mlm@helix.nih.gov