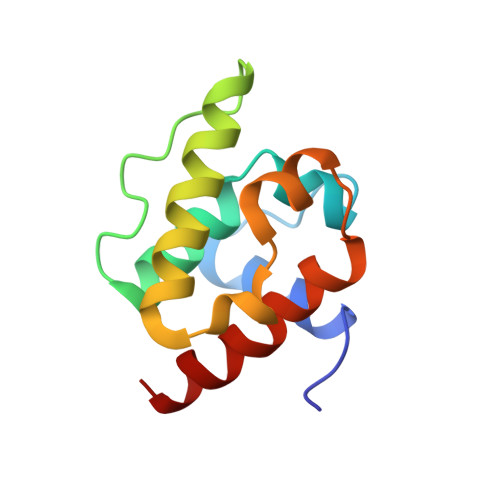
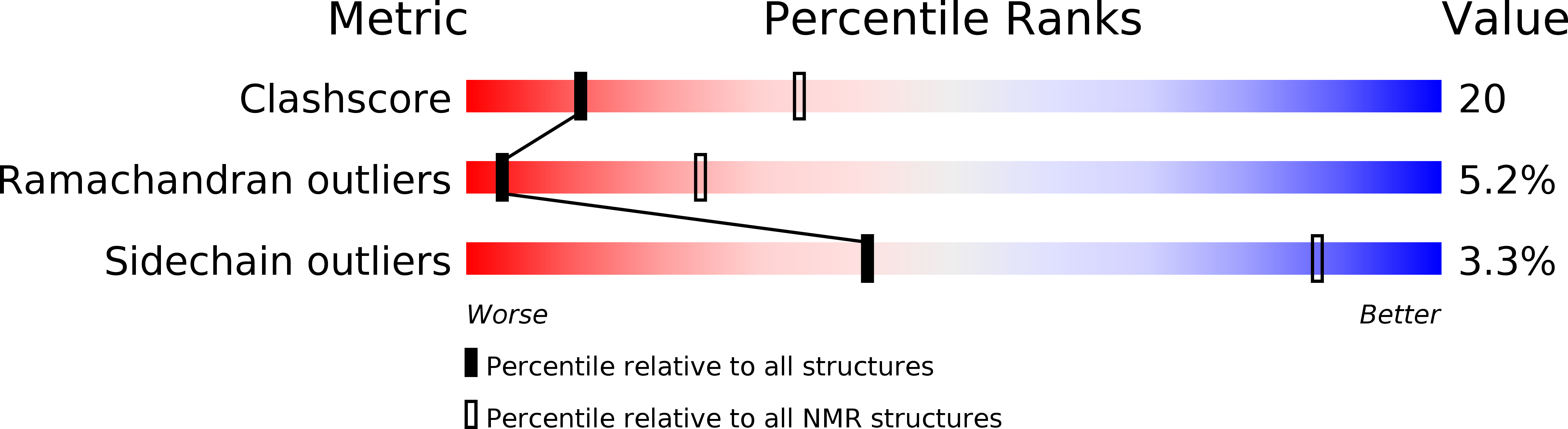Solution Structure of the Calponin Ch Domain and Fitting to the 3D-Helical Reconstruction of F-Actin:Calponin.
Bramham, J., Hodgkinson, J.L., Smith, B.O., Uhrin, D., Barlow, P.N., Winder, S.J.(2002) Structure 10: 249
- PubMed: 11839310
- DOI: https://doi.org/10.1016/s0969-2126(02)00703-7
- Primary Citation of Related Structures:
1H67 - PubMed Abstract:
Calponin is involved in the regulation of contractility and organization of the actin cytoskeleton in smooth muscle cells. It is the archetypal member of the calponin homology (CH) domain family of actin binding proteins that includes cytoskeletal linkers such as alpha-actinin, spectrin, and dystrophin, and regulatory proteins including VAV, IQGAP, and calponin. We have determined the first structure of a CH domain from a single CH domain-containing protein, that of calponin, and have fitted the NMR-derived coordinates to the 3D-helical reconstruction of the F-actin:calponin complex using cryo-electron microscopy. The tertiary fold of this single CH domain is typical of, yet significantly different from, those of the CH domains that occur in tandem pairs to form high-affinity ABDs in other proteins. We thus provide a structural insight into the mode of interaction between F-actin and CH domain-containing proteins.
Organizational Affiliation:
Department of Biochemistry, University of Leicester, Adrian Building, University Road, Leicester LE1 7RH, United Kingdom. jb139@le.ac.uk