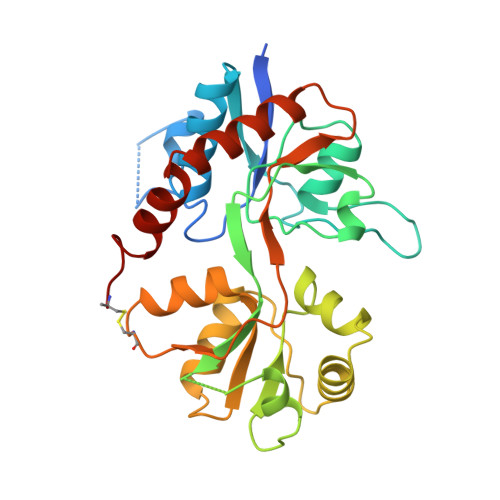
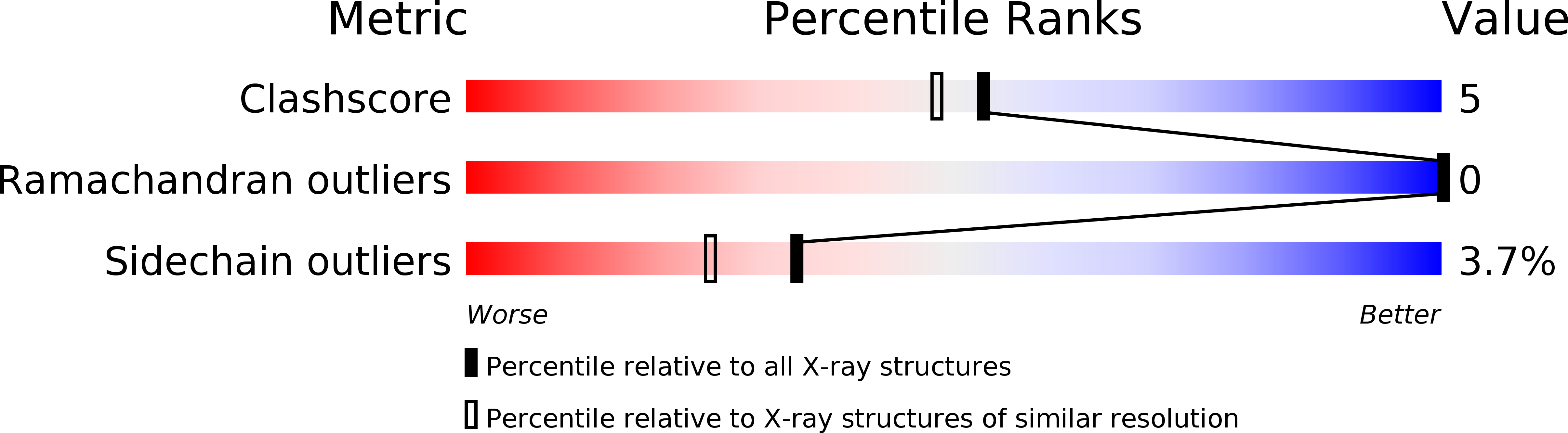Structure of a glutamate-receptor ligand-binding core in complex with kainate.
Armstrong, N., Sun, Y., Chen, G.Q., Gouaux, E.(1998) Nature 395: 913-917
- PubMed: 9804426
- DOI: https://doi.org/10.1038/27692
- Primary Citation of Related Structures:
1GR2 - PubMed Abstract:
Ionotropic glutamate receptors (iGluRs) mediate excitatory synaptic transmission in vertebrates and invertebrates through ligand-induced opening of transmembrane ion channels. iGluRs are segregated into three subtypes according to their sensitivity to the agonists AMPA (alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid), kainate (a structural analogue of glutamate) or NMDA (N-methyl-D-aspartate). iGluRs are important in the development and function of the nervous system, are essential in memory and learning, and are either implicated in or have causal roles in dysfunctions ranging from Alzheimer's, Parkinson's and Huntington's diseases, schizophrenia, epilepsy and Rasmussen's encephalitis to stroke. Development of iGluR agonists and antagonists has been hampered by a lack of high-resolution structural information. Here we describe the crystal structure of an iGluR ligand-binding region in a complex with the neurotoxin (agonist) kainate. The bilobed structure shows the determinants of receptor-agonist interactions and how ligand-binding specificity and affinity are altered by remote residues and the redox state of the conserved disulphide bond. The structure indicates mechanisms for allosteric effector action and for ligand-induced channel gating. The information provided by this structure will be essential in designing new ligands.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, New York 10032, USA.