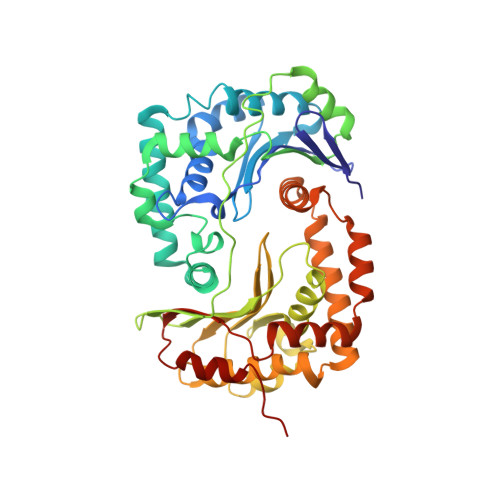
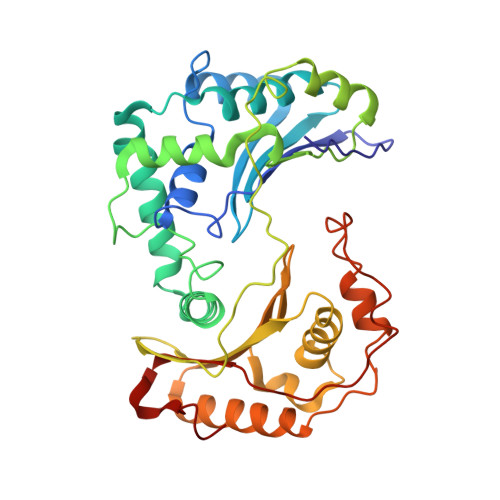
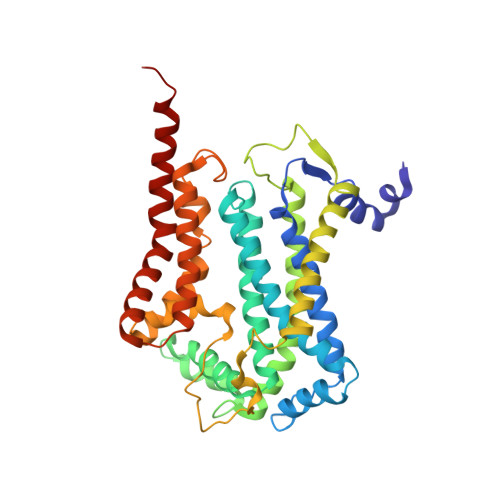
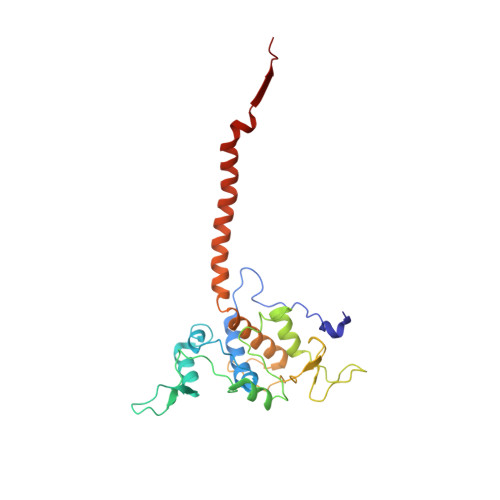
Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment.
Hunte, C., Koepke, J., Lange, C., Rossmanith, T., Michel, H.(2000) Structure 8: 669-684
- PubMed: 10873857
- DOI: https://doi.org/10.1016/s0969-2126(00)00152-0
- Primary Citation of Related Structures:
1EZV - PubMed Abstract:
The cytochrome bc(1) complex is part of the energy conversion machinery of the respiratory and photosynthetic electron transfer chains. This integral membrane protein complex catalyzes electron transfer from ubiquinol to cytochrome c. It couples the electron transfer to the electrogenic translocation of protons across the membrane via a so-called Q cycle mechanism.
Organizational Affiliation:
Max-Planck-Institut für Biophysik, Abt. Molekulare Membranbiologie, Frankfurt, 60528, Germany. hunte@mpibp-frankfurt.mpg.de.