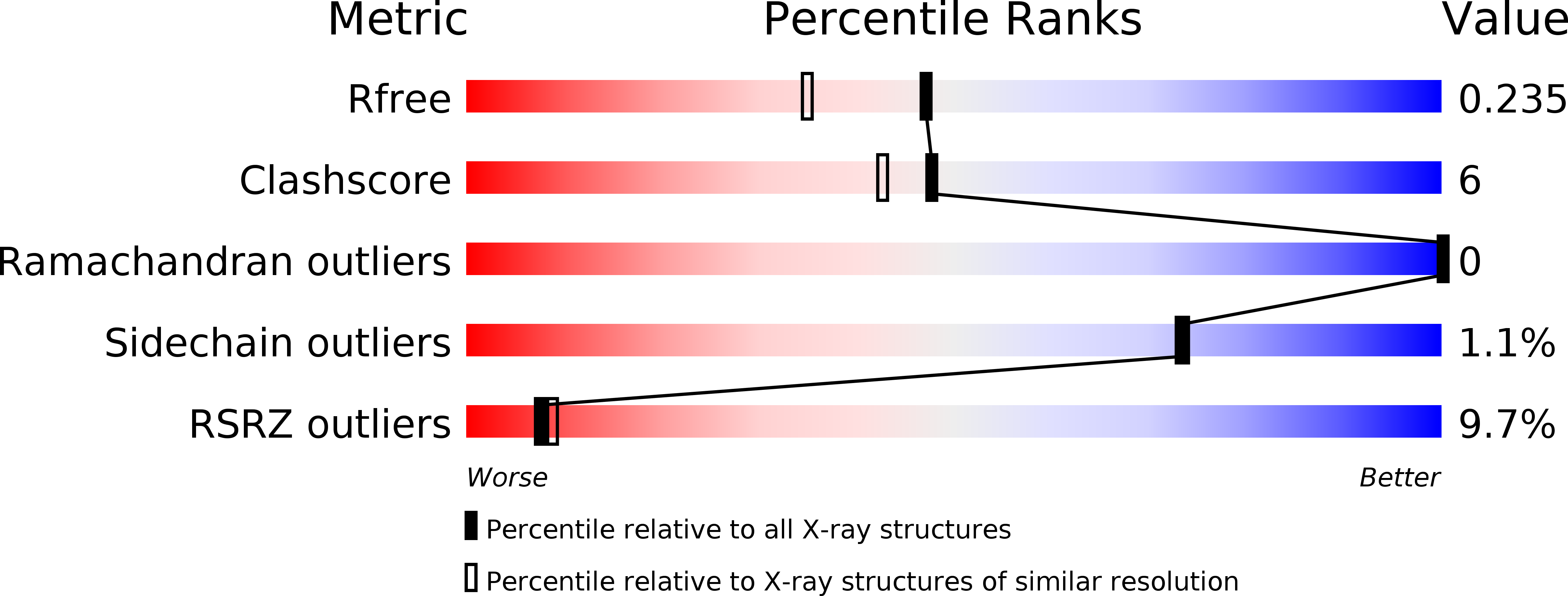
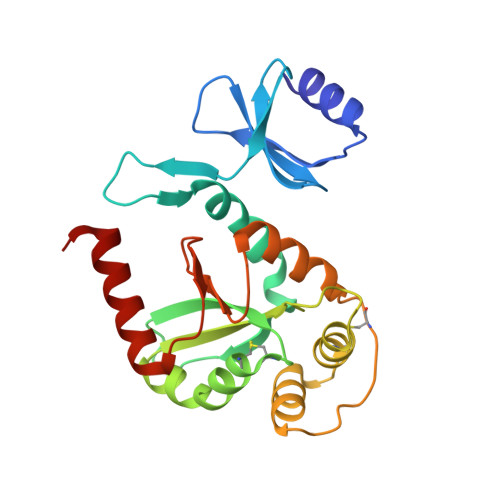
Crystal structure of the protein disulfide bond isomerase, DsbC, from Escherichia coli.
McCarthy, A.A., Haebel, P.W., Torronen, A., Rybin, V., Baker, E.N., Metcalf, P.(2000) Nat Struct Biol 7: 196-199
- PubMed: 10700276
- DOI: https://doi.org/10.1038/73295
- Primary Citation of Related Structures:
1EEJ, 1G0T - PubMed Abstract:
DsbC is one of five Escherichia coli proteins required for disulfide bond formation and is thought to function as a disulfide bond isomerase during oxidative protein folding in the periplasm. DsbC is a 2 x 23 kDa homodimer and has both protein disulfide isomerase and chaperone activity. We report the 1.9 A resolution crystal structure of oxidized DsbC where both Cys-X-X-Cys active sites form disulfide bonds. The molecule consists of separate thioredoxin-like domains joined via hinged linker helices to an N-terminal dimerization domain. The hinges allow relative movement of the active sites, and a broad uncharged cleft between them may be involved in peptide binding and DsbC foldase activities.
Organizational Affiliation:
School of Biological Sciences, Auckland University, Private Bag 92019 Auckland New Zealand.