Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family.
Nevskaya, N., Tischenko, S., Fedorov, R., Al-Karadaghi, S., Liljas, A., Kraft, A., Piendl, W., Garber, M., Nikonov, S.(2000) Structure 8: 363-371
- PubMed: 10801481
- DOI: https://doi.org/10.1016/s0969-2126(00)00116-7
- Primary Citation of Related Structures:
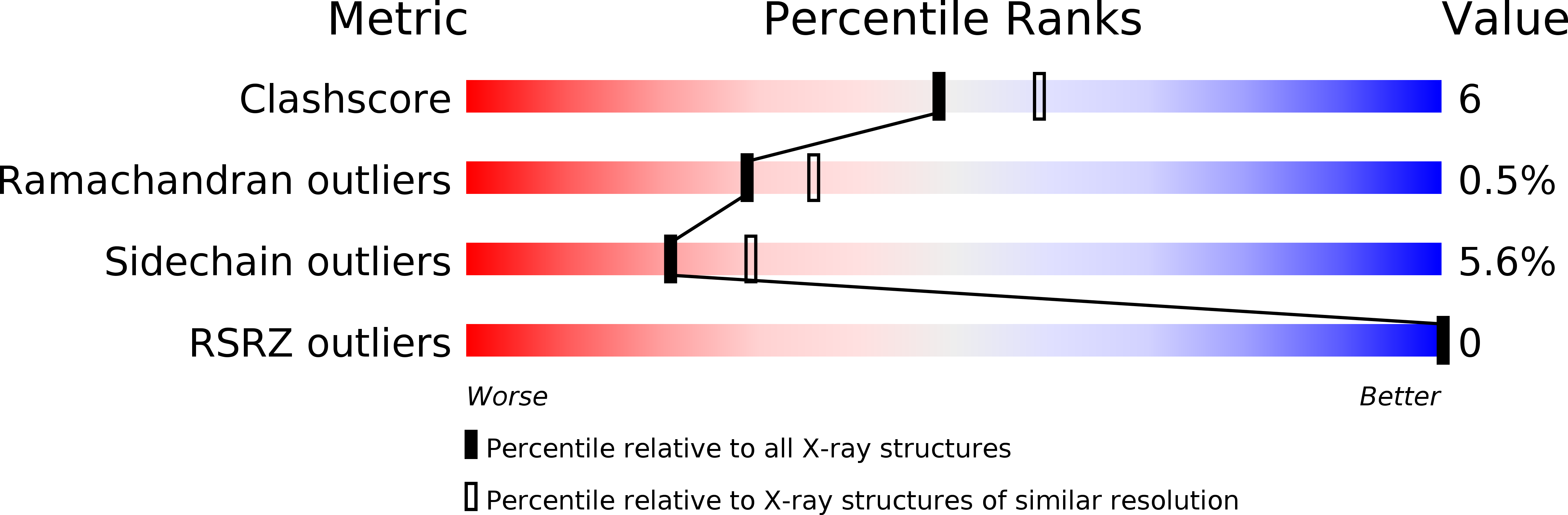
1CJS - PubMed Abstract:
L1 is an important primary rRNA-binding protein, as well as a translational repressor that binds mRNA. It was shown that L1 proteins from some bacteria and archaea are functionally interchangeable within the ribosome and in the repression of translation. The crystal structure of bacterial L1 from Thermus thermophilus (TthL1) has previously been determined. We report here the first structure of a ribosomal protein from archaea, L1 from Methanococcus jannaschii (MjaL1). The overall shape of the two-domain molecule differs dramatically from that of its bacterial counterpart (TthL1) because of the different relative orientations of the domains. Two strictly conserved regions of the amino acid sequence, each belonging to one of the domains and positioned close to each other in the interdomain cavity of TthL1, are separated by about 25 A in MjaL1 owing to a significant opening of the structure. These regions are structurally highly conserved and are proposed to be the specific RNA-binding sites. The unusually high RNA-binding affinity of MjaL1 might be explained by the exposure of its highly conserved regions. The open conformation of MjaL1 is strongly stabilized by nonconserved interdomain interactions and suggests that the closed conformations of L1 (as in TthL1) open upon RNA binding. Comparison of the two L1 protein structures reveals a high conformational variability of this ribosomal protein. Determination of the MjaL1 structure offers an additional variant for fitting the L1 protein into electron-density maps of the 50S ribosomal subunit.
Organizational Affiliation:
Institute of Protein Research, Russian Academy of Sciences, Pushchino, 142292, Moscow Region, Russia.