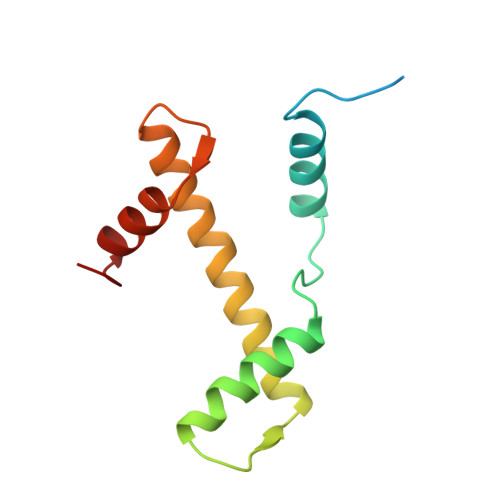
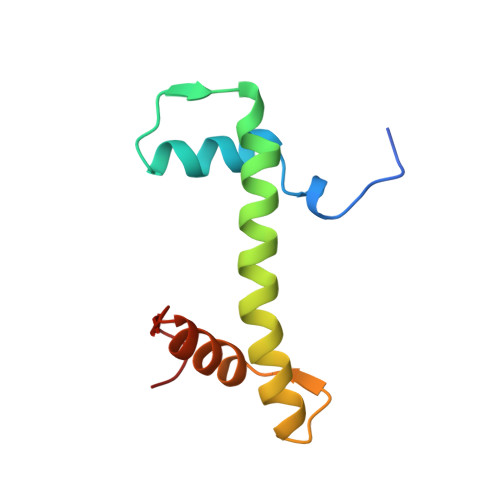
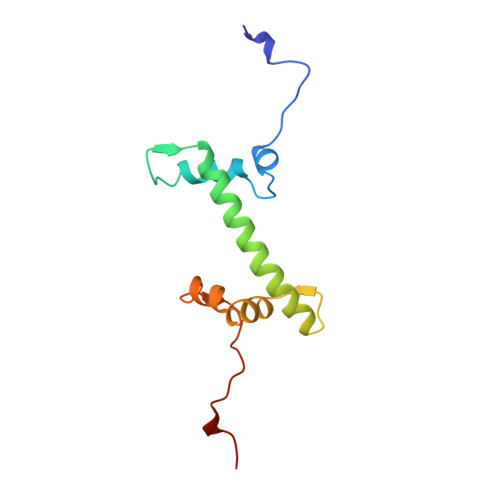
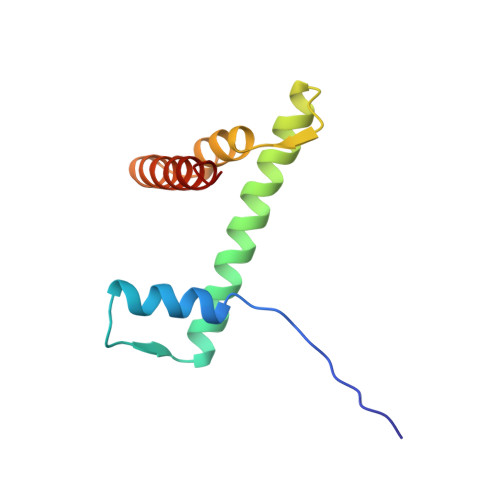
Crystal structure of the nucleosome core particle at 2.8 A resolution.
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F., Richmond, T.J.(1997) Nature 389: 251-260
- PubMed: 9305837
- DOI: https://doi.org/10.1038/38444
- Primary Citation of Related Structures:
1AOI - PubMed Abstract:
The X-ray crystal structure of the nucleosome core particle of chromatin shows in atomic detail how the histone protein octamer is assembled and how 146 base pairs of DNA are organized into a superhelix around it. Both histone/histone and histone/DNA interactions depend on the histone fold domains and additional, well ordered structure elements extending from this motif. Histone amino-terminal tails pass over and between the gyres of the DNA superhelix to contact neighbouring particles. The lack of uniformity between multiple histone/DNA-binding sites causes the DNA to deviate from ideal superhelix geometry.
Organizational Affiliation:
Institut für Molekularbiologie und Biophysik, Zürich, Switzerland.