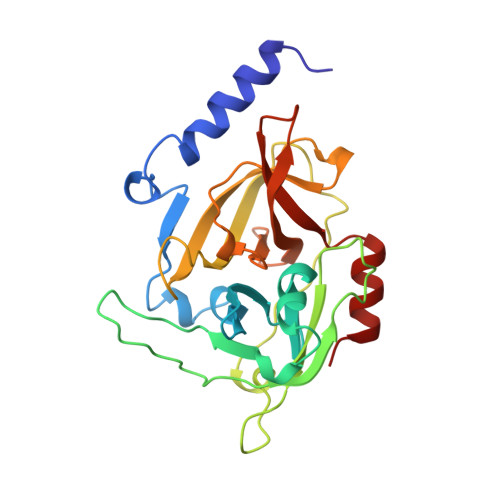
The structure of Staphylococcus aureus epidermolytic toxin A, an atypic serine protease, at 1.7 A resolution.
Cavarelli, J., Prevost, G., Bourguet, W., Moulinier, L., Chevrier, B., Delagoutte, B., Bilwes, A., Mourey, L., Rifai, S., Piemont, Y., Moras, D.(1997) Structure 5: 813-824
- PubMed: 9261066
- DOI: https://doi.org/10.1016/s0969-2126(97)00235-9
- Primary Citation of Related Structures:
1AGJ - PubMed Abstract:
Staphylococcal epidermolytic toxins A and B (ETA and ETB) are responsible for the staphylococcal scalded skin syndrome of newborn and young infants; this condition can appear just a few hours after birth. These toxins cause the disorganization and disruption of the region between the stratum spinosum and the stratum granulosum--two of the three cellular layers constituting the epidermis. The physiological substrate of ETA is not known and, consequently, its mode of action in vivo remains an unanswered question. Determination of the structure of ETA and its comparison with other serine proteases may reveal insights into ETA's catalytic mechanism. The crystal structure of staphylococcal ETA has been determined by multiple isomorphous replacement and refined at 1.7 A resolution with a crystallographic R factor of 0.184. The structure of ETA reveals it to be a new and unique member of the trypsin-like serine protease family. In contrast to other serine protease folds, ETA can be characterized by ETA-specific surface loops, a lack of cysteine bridges, an oxyanion hole which is not preformed, an S1 specific pocket designed for a negatively charged amino acid and an ETA-specific specific N-terminal helix which is shown to be crucial for substrate hydrolysis. Despite very low sequence homology between ETA and other trypsin-like serine proteases, the ETA crystal structure, together with biochemical data and site-directed mutagenesis studies, strongly confirms the classification of ETA in the Glu-endopeptidase family. Direct links can be made between the protease architecture of ETA and its biological activity.
Organizational Affiliation:
Institut de Génétique et de Biologie Moléculaire et Cellulaire, CNRS/INSERM/ULP, Illkirch, France. cava@igbmc.u-strasbg.fr