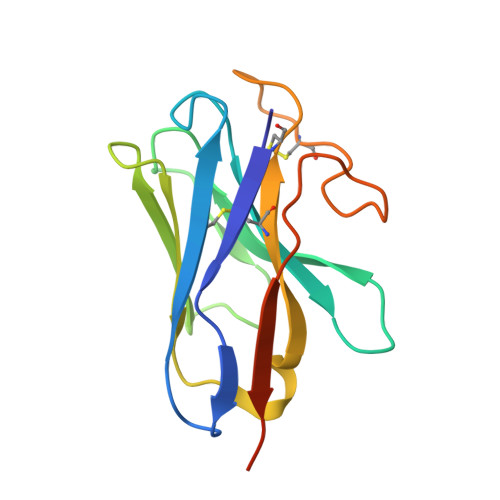
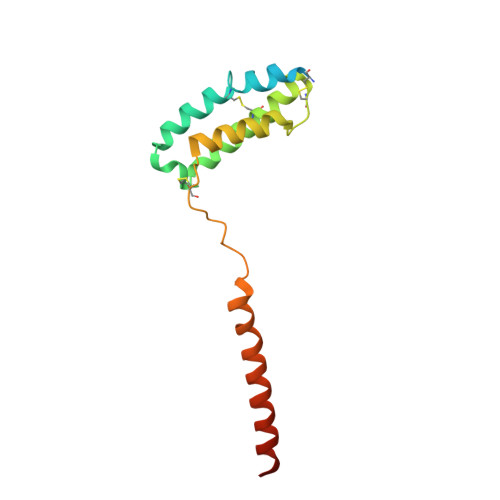
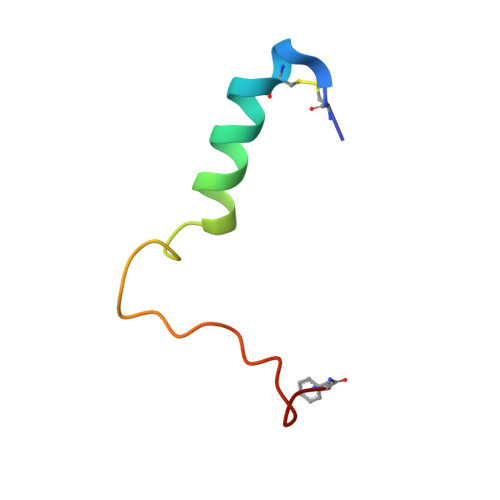
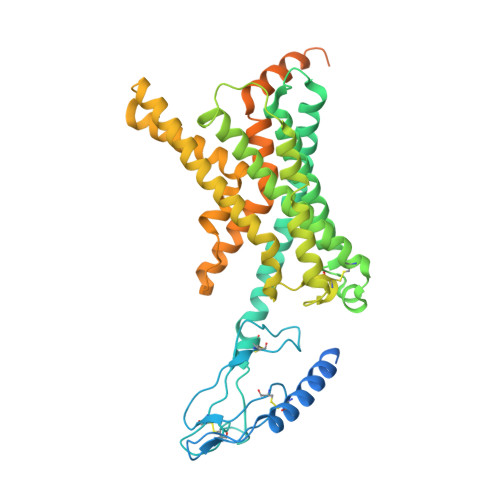
Cryo-EM Structure of the Human Amylin 1 Receptor in Complex with CGRP and Gs Protein.
Cao, J., Belousoff, M.J., Danev, R., Christopoulos, A., Wootten, D., Sexton, P.M.(2024) Biochemistry
- PubMed: 38603770
- DOI: https://doi.org/10.1021/acs.biochem.4c00114
- Primary Citation of Related Structures:
9AUC - PubMed Abstract:
Inhibition of calcitonin gene-related peptide (CGRP) or its cognate CGRP receptor (CGRPR) has arisen as a major breakthrough in the treatment of migraine. However, a second CGRP-responsive receptor exists, the amylin (Amy) 1 receptor (AMY 1 R), yet its involvement in the pathology of migraine is poorly understood. AMY 1 R and CGRPR are heterodimers consisting of receptor activity-modifying protein 1 (RAMP1) with the calcitonin receptor (CTR) and the calcitonin receptor-like receptor (CLR), respectively. Here, we present the structure of AMY 1 R in complex with CGRP and Gs protein and compare it with the reported structures of the AMY 1 R complex with rat amylin (rAmy) and the CGRPR in complex with CGRP. Despite similar protein backbones observed within the receptors and the N- and C-termini of the two peptides bound to the AMY 1 R complexes, they have distinct organization in the peptide midregions (the bypass motif) that is correlated with differences in the dynamics of the respective receptor extracellular domains. Moreover, divergent conformations of extracellular loop (ECL) 3, intracellular loop (ICL) 2, and ICL3 within the CTR and CLR protomers are evident when comparing the CGRP bound to the CGRPR and AMY 1 R, which influences the binding mode of CGRP. However, the conserved interactions made by the C-terminus of CGRP to the CGRPR and AMY 1 R are likely to account for cross-reactivity of nonpeptide CGRPR antagonists observed at AMY 1 R, which also extends to other clinically used CGRPR blockers, including antibodies.
Organizational Affiliation:
Drug Discovery Biology Theme, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, Victoria 3052, Australia.