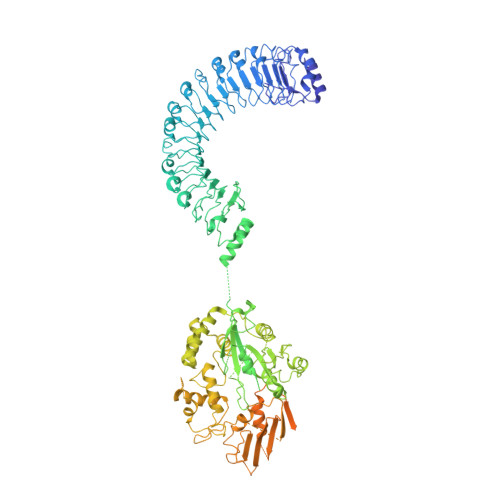Structural insights into the GTP-driven monomerization and activation of a bacterial LRRK2 homolog using allosteric nanobodies.
Galicia, C., Guaitoli, G., Fislage, M., Gloeckner, C.J., Versees, W.(2024) Elife 13
- PubMed: 38666771
- DOI: https://doi.org/10.7554/eLife.94503
- Primary Citation of Related Structures:
8R4B, 8R4C, 8R4D - PubMed Abstract:
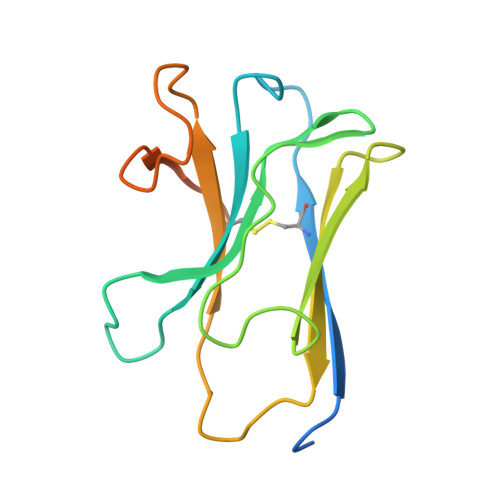
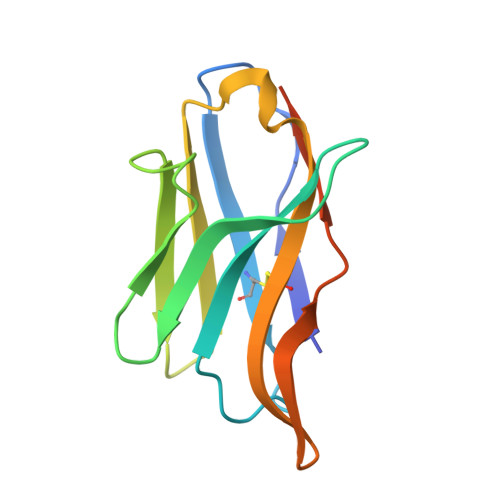
Roco proteins entered the limelight after mutations in human LRRK2 were identified as a major cause of familial Parkinson's disease. LRRK2 is a large and complex protein combining a GTPase and protein kinase activity, and disease mutations increase the kinase activity, while presumably decreasing the GTPase activity. Although a cross-communication between both catalytic activities has been suggested, the underlying mechanisms and the regulatory role of the GTPase domain remain unknown. Several structures of LRRK2 have been reported, but structures of Roco proteins in their activated GTP-bound state are lacking. Here, we use single-particle cryo-electron microscopy to solve the structure of a bacterial Roco protein (CtRoco) in its GTP-bound state, aided by two conformation-specific nanobodies: Nb Roco1 and Nb Roco2 . This structure presents CtRoco in an active monomeric state, featuring a very large GTP-induced conformational change using the LRR-Roc linker as a hinge. Furthermore, this structure shows how Nb Roco1 and Nb Roco2 collaborate to activate CtRoco in an allosteric way. Altogether, our data provide important new insights into the activation mechanism of Roco proteins, with relevance to LRRK2 regulation, and suggest new routes for the allosteric modulation of their GTPase activity.
Organizational Affiliation:
Structural Biology Brussels, Vrije Universiteit Brussel, Brussels, Belgium.