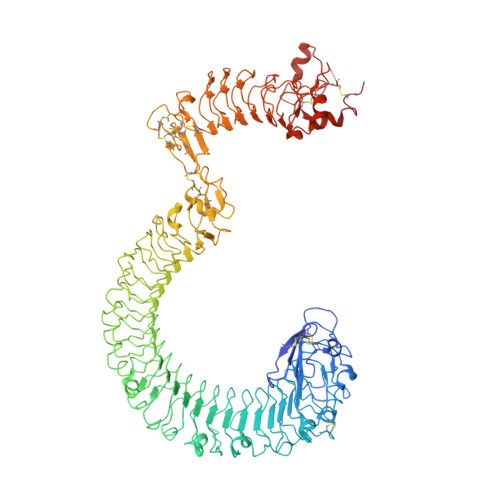
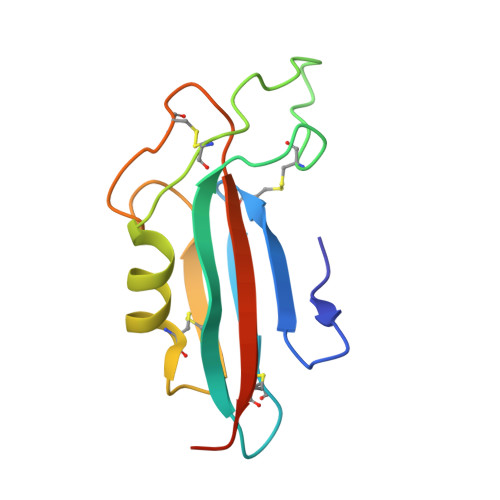
Structural basis and functional roles for Toll-like receptor binding to Latrophilin adhesion-GPCR in embryo development
Carmona Rosas, G., Li, J., Smith, J.S., Cheng, S., Baltrusaitis, E., Nawrocka, W.I., Zhao, M., Kratsios, P., Arac, D., Ozkan, E.To be published.