Hidden protein disorder: Deciphering the structural organisation and dynamics of a non-canonical CP12 from the diatom Thalassiosira pseudonana
Bonucci, A., Wang, T., Baroudi, H., Etienne, E., Gerbaud, G., Mileo, E., Parsiegla, G., Yamato, T., Gontero, B., Launay, H., Belle, V., Receveur-Brechot, V.(2025) Febs J
- PubMed: 40674273
- DOI: https://doi.org/10.1111/febs.70187
- Primary Citation of Related Structures:
9A9R - PubMed Abstract:
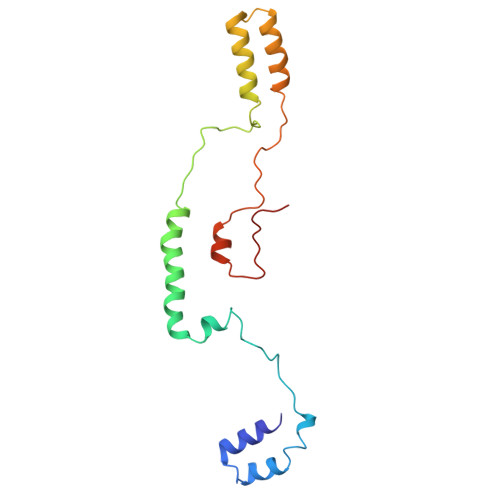
The chloroplastic protein CP12 from the diatom Thalassiosira pseudonana exhibits atypical and enigmatic structural properties that have so far hindered our understanding of its functions. Here, we used AlphaFold to generate a three-dimensional (3D) model of the structure of the protein. However, this model did not accurately describe the small-angle X-ray scattering (SAXS) data previously obtained. We have therefore undertaken a study using site-directed spin labelling combined with electron paramagnetic resonance (SDSL-EPR) to characterise the structural dynamics of this atypical CP12 and to investigate its dimeric organisation using double electron-electron resonance (DEER). We then performed molecular dynamics (MD) simulations, constrained by SAXS and DEER data, to refine the AlphaFold model and take into account the flexibility and disordered propensities of this protein. The combination of the experimental techniques together with the in silico AlphaFold and MD simulations reveals that the dimer is organised in an antiparallel arrangement of each monomer and that the C-terminal regions are highly flexible and partly disordered. Additionally, this diatom CP12 contains four structured domains likely to bind phosphoribulokinase regardless of their redox state. Our structural data therefore provide insights into the function of this protein in the regulation of the Calvin cycle and of photosynthesis in diatoms, whereas its structural organisation is completely different from any of its homologous counterparts from Plantae and cyanobacteria.
- CNRS, BIP UMR7281, Aix Marseille Univ, France.
Organizational Affiliation: